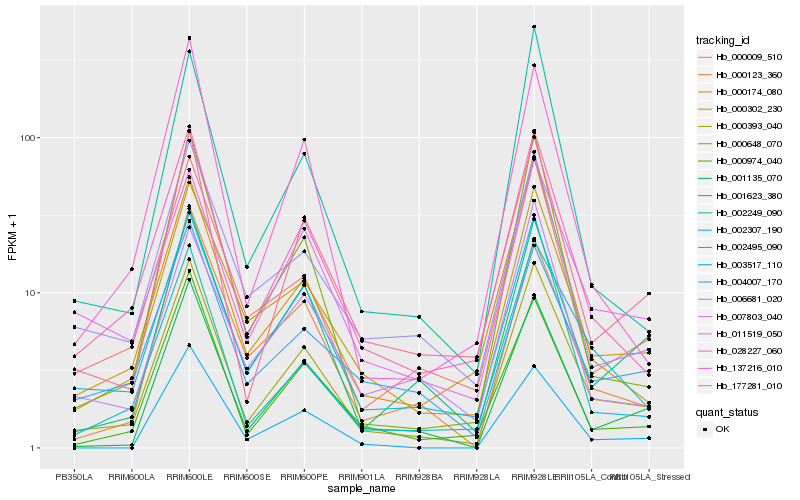
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000393_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 2 |
Hb_137216_010 |
0.0914021918 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 3 |
Hb_000302_230 |
0.0959022861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_177281_010 |
0.1020481095 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 5 |
Hb_000009_510 |
0.1099256113 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 6 |
Hb_001135_070 |
0.1103709853 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 7 |
Hb_000974_040 |
0.1130337588 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 8 |
Hb_000648_070 |
0.114250926 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 9 |
Hb_002307_190 |
0.1185635752 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 10 |
Hb_002495_090 |
0.1199520072 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000174_080 |
0.1220250935 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 12 |
Hb_001623_380 |
0.1223122996 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002249_090 |
0.1292514843 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_028227_060 |
0.1311616913 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 15 |
Hb_011519_050 |
0.1318914707 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |
| 16 |
Hb_004007_170 |
0.135574174 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_006681_020 |
0.1364196067 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000123_360 |
0.1366472812 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_003517_110 |
0.138428063 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 20 |
Hb_007803_040 |
0.1442858553 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |