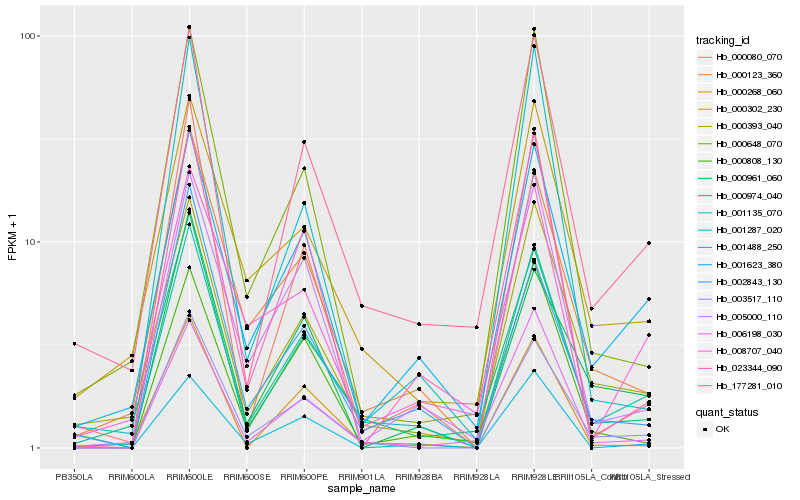
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001135_070 |
0.0 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 2 |
Hb_003517_110 |
0.1022347404 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 3 |
Hb_177281_010 |
0.1046392885 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 4 |
Hb_001623_380 |
0.106620474 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000974_040 |
0.1095392505 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 6 |
Hb_000393_040 |
0.1103709853 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 7 |
Hb_000123_360 |
0.1200465586 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_005000_110 |
0.1307377163 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000080_070 |
0.139073744 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000302_230 |
0.1408149602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000268_060 |
0.1452850722 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_006198_030 |
0.146284563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000648_070 |
0.146771224 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 14 |
Hb_023344_090 |
0.1498502593 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 15 |
Hb_000808_130 |
0.1505053227 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 16 |
Hb_001488_250 |
0.151717175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008707_040 |
0.1539566907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000961_060 |
0.1542069398 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 19 |
Hb_001287_020 |
0.154698898 |
- |
- |
SulA [Manihot esculenta] |
| 20 |
Hb_002843_130 |
0.1547321099 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |