| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
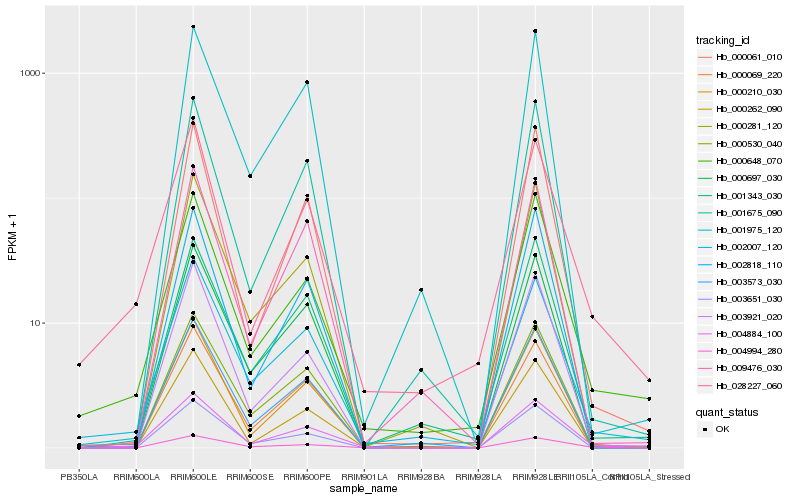
Hb_004884_100 |
0.0 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 2 |
Hb_004994_280 |
0.091991096 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 3 |
Hb_002007_120 |
0.1002648671 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 4 |
Hb_028227_060 |
0.1049793403 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 5 |
Hb_000648_070 |
0.1058179287 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 6 |
Hb_000530_040 |
0.1136491768 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003573_030 |
0.116001355 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 8 |
Hb_000061_010 |
0.1176304029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000069_220 |
0.118397883 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 10 |
Hb_001675_090 |
0.1197663898 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000210_030 |
0.1207429559 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 12 |
Hb_000697_030 |
0.1211399236 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 13 |
Hb_001343_030 |
0.1212914651 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000281_120 |
0.1219014294 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002818_110 |
0.1221403621 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 16 |
Hb_000262_090 |
0.123059219 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 17 |
Hb_003921_020 |
0.1258300335 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 18 |
Hb_001975_120 |
0.1263277803 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 19 |
Hb_009476_030 |
0.1265259827 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 20 |
Hb_003651_030 |
0.126702471 |
- |
- |
lyase, putative [Ricinus communis] |