| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
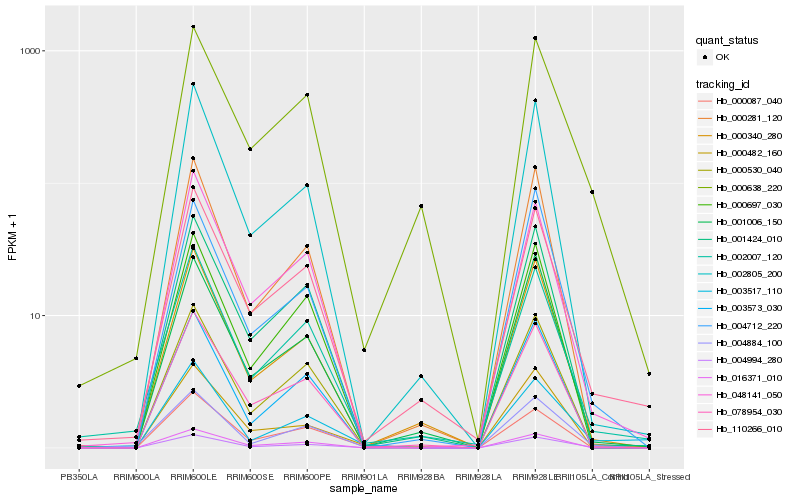
Hb_004994_280 |
0.0 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 2 |
Hb_004884_100 |
0.091991096 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 3 |
Hb_078954_030 |
0.1130497461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000530_040 |
0.1135122843 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_110266_010 |
0.1150741262 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 6 |
Hb_048141_050 |
0.115565455 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 7 |
Hb_001424_010 |
0.1171077924 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000087_040 |
0.1185256669 |
- |
- |
Autoinhibited Ca(2+)-ATPase 10 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000340_280 |
0.1186832689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000638_220 |
0.1187251717 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002007_120 |
0.1190801905 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003517_110 |
0.1219694512 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 13 |
Hb_000281_120 |
0.1232401396 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000482_160 |
0.1241270976 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 15 |
Hb_016371_010 |
0.1246230375 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 16 |
Hb_000697_030 |
0.1246294536 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 17 |
Hb_002805_200 |
0.1253504629 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 18 |
Hb_003573_030 |
0.126069426 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 19 |
Hb_004712_220 |
0.1271480031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001006_150 |
0.1275361335 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |