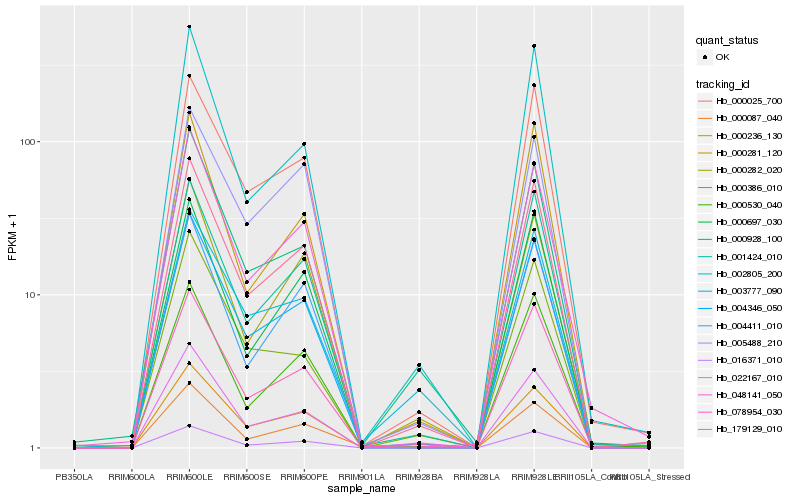
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004346_050 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 2 |
Hb_179129_010 |
0.0343374994 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 3 |
Hb_016371_010 |
0.0374288284 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_078954_030 |
0.0448120045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000025_700 |
0.0573485373 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 6 |
Hb_001424_010 |
0.0578131525 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_048141_050 |
0.0624859116 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 8 |
Hb_022167_010 |
0.0728284509 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 9 |
Hb_000087_040 |
0.0750264711 |
- |
- |
Autoinhibited Ca(2+)-ATPase 10 isoform 1 [Theobroma cacao] |
| 10 |
Hb_004411_010 |
0.0756163279 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 11 |
Hb_000236_130 |
0.0771671027 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 12 |
Hb_000530_040 |
0.0776335165 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000928_100 |
0.0816195468 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 14 |
Hb_000697_030 |
0.0821302539 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 15 |
Hb_002805_200 |
0.0833583252 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 16 |
Hb_000282_020 |
0.0843110246 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005488_210 |
0.0874222975 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 18 |
Hb_003777_090 |
0.0923677097 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 19 |
Hb_000386_010 |
0.0929118367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000281_120 |
0.093747805 |
- |
- |
protein binding protein, putative [Ricinus communis] |