| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
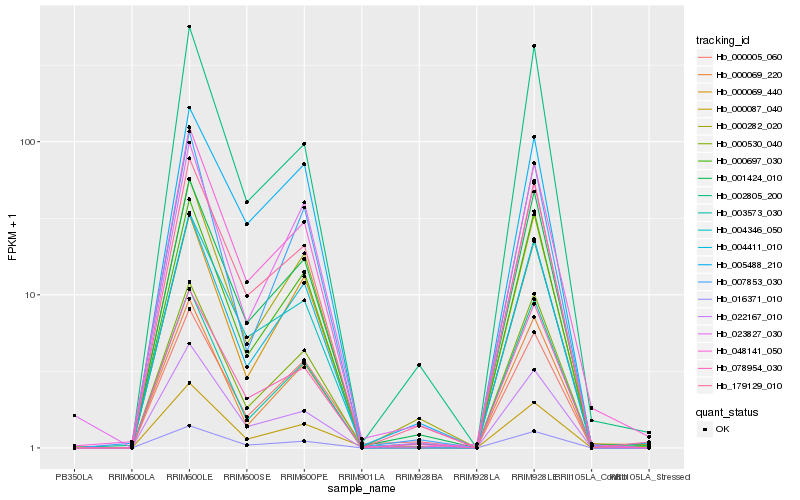
Hb_000087_040 |
0.0 |
- |
- |
Autoinhibited Ca(2+)-ATPase 10 isoform 1 [Theobroma cacao] |
| 2 |
Hb_016371_010 |
0.0732205759 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 3 |
Hb_004411_010 |
0.0733382446 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 4 |
Hb_004346_050 |
0.0750264711 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 5 |
Hb_048141_050 |
0.0797992774 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 6 |
Hb_000005_060 |
0.0824151017 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 7 |
Hb_179129_010 |
0.0840208548 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 8 |
Hb_000282_020 |
0.0885238185 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000069_220 |
0.0898977577 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 10 |
Hb_000530_040 |
0.0913541823 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001424_010 |
0.0919999749 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000697_030 |
0.0921879123 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 13 |
Hb_078954_030 |
0.0924332394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000069_440 |
0.0953332953 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 15 |
Hb_007853_030 |
0.1020969509 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 16 |
Hb_022167_010 |
0.1054409146 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 17 |
Hb_003573_030 |
0.1055889254 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 18 |
Hb_023827_030 |
0.1079655105 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_002805_200 |
0.1104087417 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 20 |
Hb_005488_210 |
0.1114913956 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |