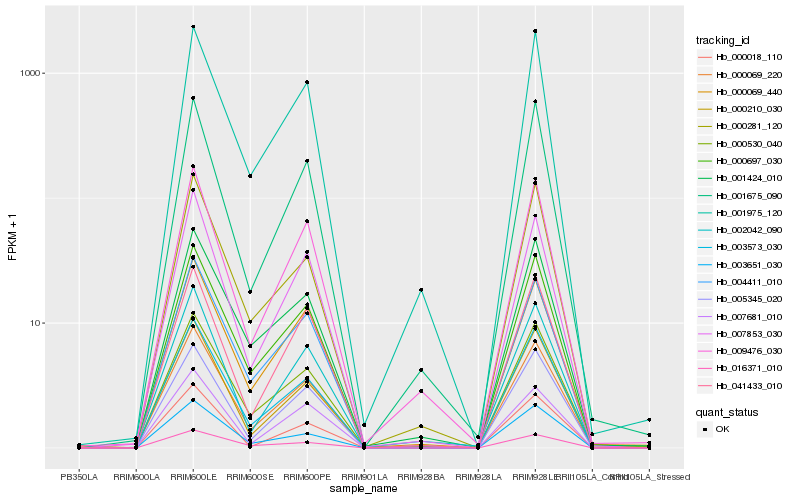
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_220 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF023 [Jatropha curcas] |
| 2 |
Hb_000697_030 |
0.0354972254 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Pyrus x bretschneideri] |
| 3 |
Hb_003573_030 |
0.0422040713 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 4 |
Hb_004411_010 |
0.0427267034 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |
| 5 |
Hb_007853_030 |
0.0501637629 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 6 |
Hb_000530_040 |
0.0545603815 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000210_030 |
0.0612919227 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 8 |
Hb_000069_440 |
0.061417535 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 9 |
Hb_001975_120 |
0.062347511 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 10 |
Hb_009476_030 |
0.0653687445 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 11 |
Hb_005345_020 |
0.0684065348 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 12 |
Hb_007681_010 |
0.0687703202 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 13 |
Hb_016371_010 |
0.0689794118 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 14 |
Hb_041433_010 |
0.0704425208 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_002042_090 |
0.0704938347 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 16 |
Hb_001424_010 |
0.0705564004 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_001675_090 |
0.0728482312 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000281_120 |
0.0741981391 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_000018_110 |
0.0746728549 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 20 |
Hb_003651_030 |
0.0756467164 |
- |
- |
lyase, putative [Ricinus communis] |