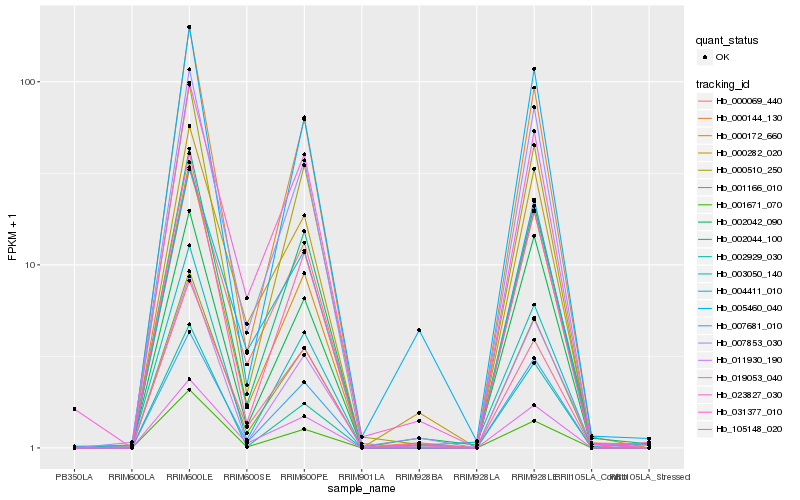
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001166_010 |
0.0 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 2 |
Hb_007853_030 |
0.0473842632 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 3 |
Hb_000144_130 |
0.051005552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002044_100 |
0.0556379948 |
- |
- |
PREDICTED: uncharacterized protein LOC105643555 [Jatropha curcas] |
| 5 |
Hb_031377_010 |
0.058819339 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 6 |
Hb_000510_250 |
0.0598278219 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 7 |
Hb_019053_040 |
0.060276022 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_007681_010 |
0.0610898256 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 9 |
Hb_105148_020 |
0.0646989896 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 10 |
Hb_000172_660 |
0.0659267136 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_000282_020 |
0.0686282534 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001671_070 |
0.0707087528 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_005460_040 |
0.0738172107 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 14 |
Hb_000069_440 |
0.0763690427 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 15 |
Hb_023827_030 |
0.0769529924 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_011930_190 |
0.0771675547 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002042_090 |
0.0792855676 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 18 |
Hb_003050_140 |
0.0842363072 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 19 |
Hb_002929_030 |
0.084601025 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 20 |
Hb_004411_010 |
0.0852801147 |
- |
- |
PREDICTED: NADP-dependent alkenal double bond reductase P2-like isoform X5 [Gossypium raimondii] |