| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
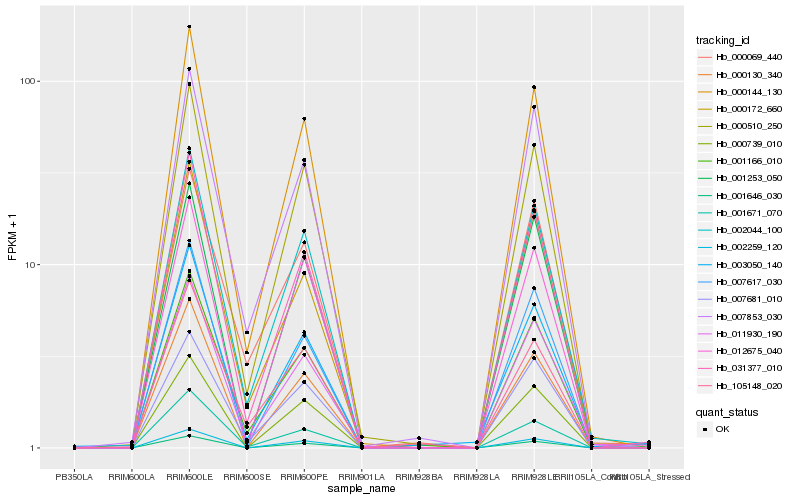
Hb_002044_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643555 [Jatropha curcas] |
| 2 |
Hb_000144_130 |
0.0326643503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000510_250 |
0.0483720915 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 4 |
Hb_031377_010 |
0.0511898177 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 5 |
Hb_001166_010 |
0.0556379948 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 6 |
Hb_011930_190 |
0.0600193512 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007681_010 |
0.0630296354 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 8 |
Hb_001671_070 |
0.0671145544 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_007853_030 |
0.0692861998 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 10 |
Hb_000172_660 |
0.0702484135 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_002259_120 |
0.0774823516 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_003050_140 |
0.0779844917 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 13 |
Hb_001646_030 |
0.0798511301 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 14 |
Hb_000739_010 |
0.0808317443 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 15 |
Hb_012675_040 |
0.0812287192 |
- |
- |
PREDICTED: uncharacterized protein LOC105647773 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000130_340 |
0.0827872399 |
- |
- |
- |
| 17 |
Hb_105148_020 |
0.0834780457 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 18 |
Hb_000069_440 |
0.0878705339 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 19 |
Hb_001253_050 |
0.0883350472 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 20 |
Hb_007617_030 |
0.0897230301 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |