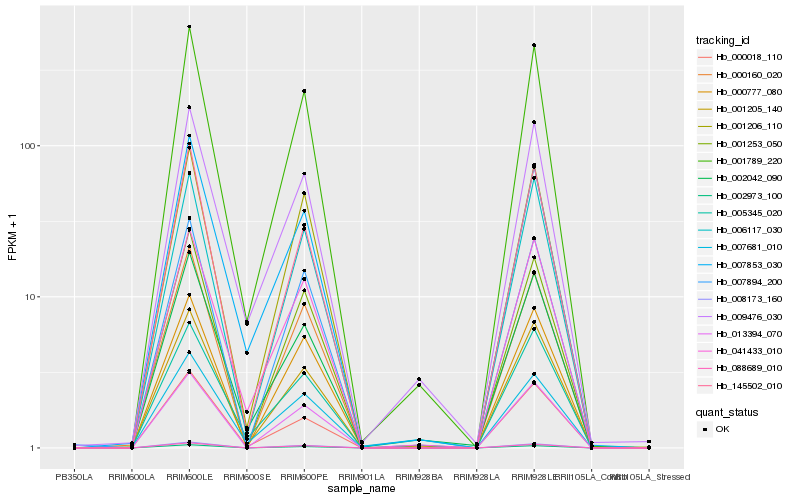
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_220 |
0.0 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 2 |
Hb_001205_140 |
0.033380543 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 3 |
Hb_041433_010 |
0.0472083821 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_006117_030 |
0.0483588963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002042_090 |
0.0509743588 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 6 |
Hb_005345_020 |
0.0536009977 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 7 |
Hb_007681_010 |
0.0551094078 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 8 |
Hb_145502_010 |
0.0553432659 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 9 |
Hb_001206_110 |
0.0567686067 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 10 |
Hb_013394_070 |
0.0575500167 |
- |
- |
hypothetical protein POPTR_0013s12090g [Populus trichocarpa] |
| 11 |
Hb_001253_050 |
0.0579233464 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 12 |
Hb_009476_030 |
0.0583977427 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 13 |
Hb_002973_100 |
0.0597448334 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Populus euphratica] |
| 14 |
Hb_000018_110 |
0.0598475405 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 15 |
Hb_008173_160 |
0.062101848 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 16 |
Hb_088689_010 |
0.0621664488 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 17 |
Hb_007853_030 |
0.0624895803 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 18 |
Hb_007894_200 |
0.0632700162 |
- |
- |
ubiquitin-conjugating enzyme variant, putative [Ricinus communis] |
| 19 |
Hb_000160_020 |
0.0648325917 |
- |
- |
PREDICTED: gamma-interferon-inducible lysosomal thiol reductase-like [Jatropha curcas] |
| 20 |
Hb_000777_080 |
0.0658588564 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |