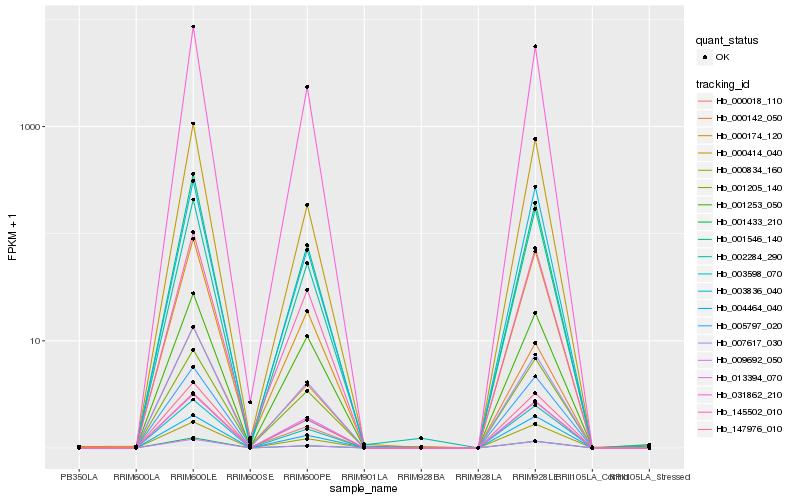
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147976_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_031862_210 |
0.0222602306 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 3 |
Hb_003598_070 |
0.0236982328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009692_050 |
0.0253263324 |
- |
- |
- |
| 5 |
Hb_145502_010 |
0.0262413433 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 6 |
Hb_002284_290 |
0.0368708283 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 7 |
Hb_000834_160 |
0.0411547252 |
- |
- |
hypothetical protein JCGZ_07239 [Jatropha curcas] |
| 8 |
Hb_001433_210 |
0.0420075354 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 9 |
Hb_000018_110 |
0.0469954932 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 10 |
Hb_000174_120 |
0.0515421107 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_001205_140 |
0.0537462545 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 12 |
Hb_003836_040 |
0.0552963007 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 13 |
Hb_000142_050 |
0.0577631251 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000414_040 |
0.0604028437 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 15 |
Hb_001546_140 |
0.0622036114 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 16 |
Hb_005797_020 |
0.0627409823 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 17 |
Hb_001253_050 |
0.0646450084 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 18 |
Hb_013394_070 |
0.0658242857 |
- |
- |
hypothetical protein POPTR_0013s12090g [Populus trichocarpa] |
| 19 |
Hb_007617_030 |
0.0670910406 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 20 |
Hb_004464_040 |
0.067423998 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |