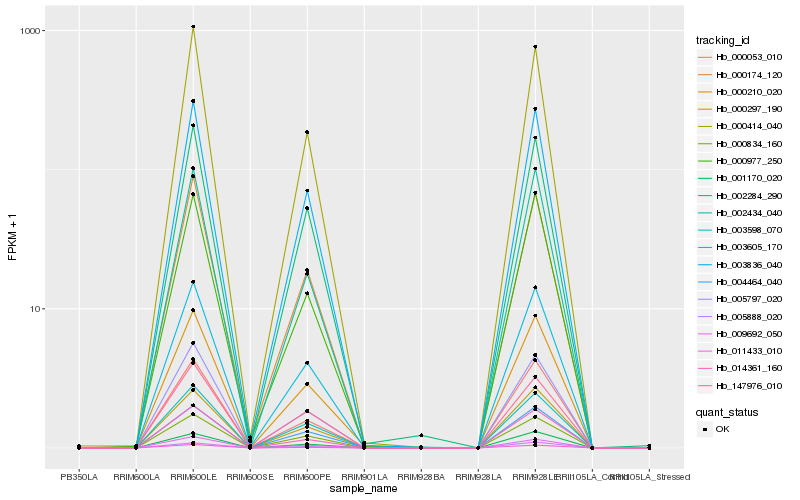
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003836_040 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 2 |
Hb_003605_170 |
0.018544813 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 3 |
Hb_002284_290 |
0.0298122338 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 4 |
Hb_000210_020 |
0.0318927142 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |
| 5 |
Hb_000174_120 |
0.0350321187 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 6 |
Hb_005797_020 |
0.0358149886 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 7 |
Hb_000834_160 |
0.0372187617 |
- |
- |
hypothetical protein JCGZ_07239 [Jatropha curcas] |
| 8 |
Hb_003598_070 |
0.0378022489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009692_050 |
0.0383764988 |
- |
- |
- |
| 10 |
Hb_000297_190 |
0.0408531458 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 11 |
Hb_011433_010 |
0.0437662338 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 12 |
Hb_000053_010 |
0.0462299764 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 13 |
Hb_000414_040 |
0.0464575409 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 14 |
Hb_000977_250 |
0.0497221177 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_014361_160 |
0.0515236354 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 16 |
Hb_002434_040 |
0.0519176974 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 17 |
Hb_147976_010 |
0.0552963007 |
- |
- |
- |
| 18 |
Hb_001170_020 |
0.0555500491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004464_040 |
0.0558226839 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 20 |
Hb_005888_020 |
0.0565476635 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |