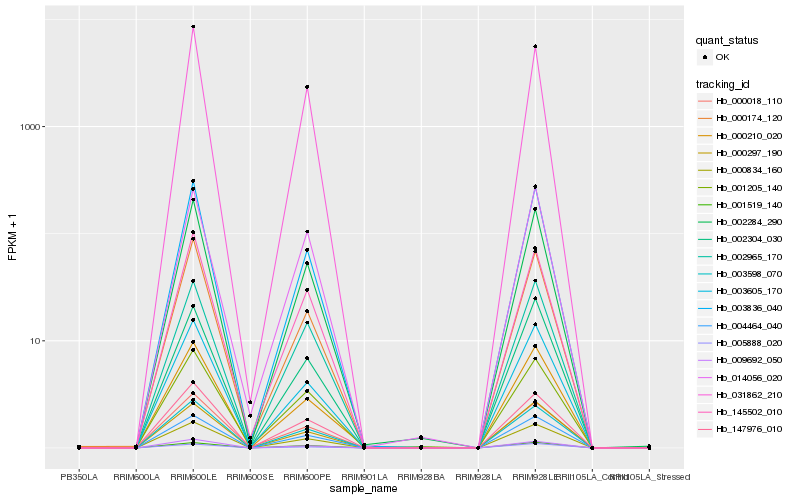
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_07239 [Jatropha curcas] |
| 2 |
Hb_003598_070 |
0.0182344334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002284_290 |
0.0315777642 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 4 |
Hb_003836_040 |
0.0372187617 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 5 |
Hb_147976_010 |
0.0411547252 |
- |
- |
- |
| 6 |
Hb_004464_040 |
0.0412142293 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 7 |
Hb_009692_050 |
0.0429430531 |
- |
- |
- |
| 8 |
Hb_002965_170 |
0.0435172751 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 9 |
Hb_000297_190 |
0.0455116613 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 10 |
Hb_003605_170 |
0.0492869166 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 11 |
Hb_000210_020 |
0.0531333124 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |
| 12 |
Hb_145502_010 |
0.0534242957 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 13 |
Hb_001205_140 |
0.055317313 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 14 |
Hb_014056_020 |
0.0565673154 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000018_110 |
0.0576061866 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 16 |
Hb_000174_120 |
0.057669316 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 17 |
Hb_005888_020 |
0.0603946577 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 18 |
Hb_002304_030 |
0.0608381181 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 19 |
Hb_001519_140 |
0.0611758981 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 20 |
Hb_031862_210 |
0.0620769922 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |