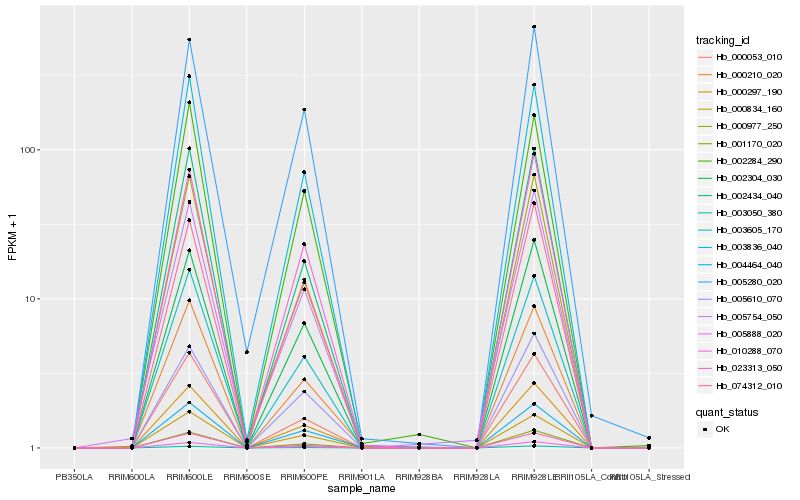
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000297_190 |
0.0 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 2 |
Hb_005888_020 |
0.0167221772 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 3 |
Hb_001170_020 |
0.0232929571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002304_030 |
0.0258379219 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 5 |
Hb_010288_070 |
0.0403688225 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 6 |
Hb_003836_040 |
0.0408531458 |
- |
- |
PREDICTED: cytochrome P450 77A3-like [Jatropha curcas] |
| 7 |
Hb_003050_380 |
0.0417142145 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 8 |
Hb_003605_170 |
0.0422781223 |
- |
- |
hypothetical protein POPTR_0013s12020g [Populus trichocarpa] |
| 9 |
Hb_000834_160 |
0.0455116613 |
- |
- |
hypothetical protein JCGZ_07239 [Jatropha curcas] |
| 10 |
Hb_000210_020 |
0.0473816328 |
transcription factor |
TF Family: OFP |
Ovate family protein 13, putative [Theobroma cacao] |
| 11 |
Hb_000977_250 |
0.0488922472 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 12 |
Hb_000053_010 |
0.0489793972 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 13 |
Hb_005610_070 |
0.0497542878 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 14 |
Hb_002434_040 |
0.0519399856 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 15 |
Hb_004464_040 |
0.0537092478 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 16 |
Hb_005754_050 |
0.0538829323 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 17 |
Hb_023313_050 |
0.0546058245 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 18 |
Hb_005280_020 |
0.0550863349 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 19 |
Hb_074312_010 |
0.0582366108 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 20 |
Hb_002284_290 |
0.0583388692 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |