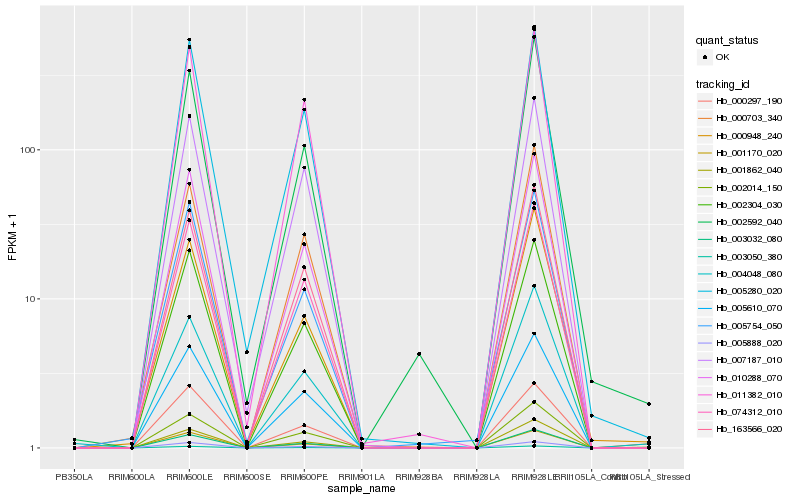
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003032_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002014_150 |
0.0279443934 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_010288_070 |
0.0281899262 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 4 |
Hb_001862_040 |
0.031885899 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 5 |
Hb_163566_020 |
0.0321028636 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_005610_070 |
0.0373145015 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 7 |
Hb_074312_010 |
0.040380377 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 8 |
Hb_003050_380 |
0.042262709 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 9 |
Hb_002304_030 |
0.0427199433 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 10 |
Hb_005888_020 |
0.0461341644 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 11 |
Hb_001170_020 |
0.0541345704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005280_020 |
0.0559583043 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 13 |
Hb_000703_340 |
0.0580439594 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_011382_010 |
0.0598323911 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 15 |
Hb_000297_190 |
0.062001688 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 16 |
Hb_000948_240 |
0.0627456578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005754_050 |
0.0637758599 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 18 |
Hb_007187_010 |
0.0638748369 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 19 |
Hb_004048_080 |
0.0664323821 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 20 |
Hb_002592_040 |
0.0681892763 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |