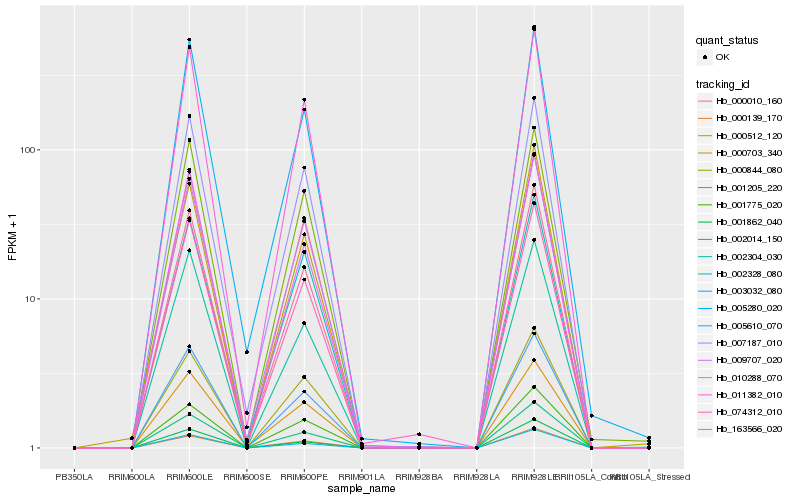
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163566_020 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_002014_150 |
0.0156075107 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_074312_010 |
0.0253589767 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 4 |
Hb_003032_080 |
0.0321028636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005610_070 |
0.0338374505 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 6 |
Hb_011382_010 |
0.0379921596 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 7 |
Hb_007187_010 |
0.0381362081 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 8 |
Hb_010288_070 |
0.0413141865 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 9 |
Hb_009707_020 |
0.0418124892 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 10 |
Hb_000010_160 |
0.0476917809 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 11 |
Hb_000703_340 |
0.0486814689 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000139_170 |
0.0488664496 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 13 |
Hb_005280_020 |
0.0494570883 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 14 |
Hb_001775_020 |
0.0513804412 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 15 |
Hb_002304_030 |
0.0544794038 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 16 |
Hb_001862_040 |
0.0547391745 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 17 |
Hb_000844_080 |
0.0554935158 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 18 |
Hb_001205_220 |
0.0577696579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000512_120 |
0.058548191 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 20 |
Hb_002328_080 |
0.0624029711 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |