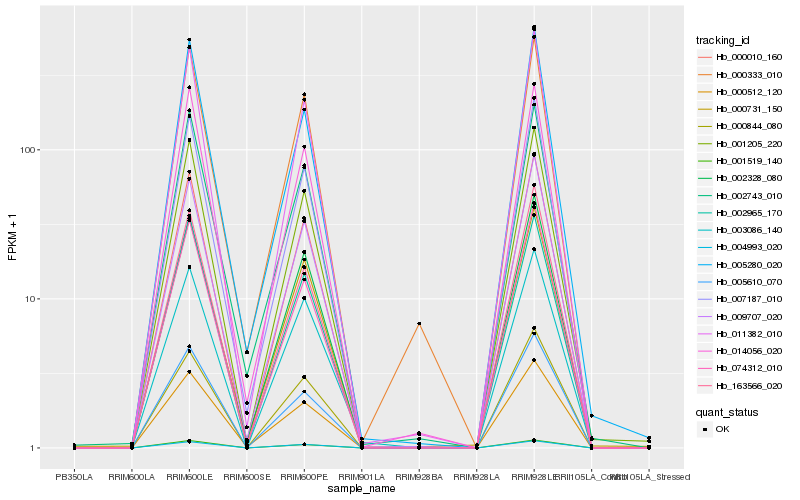
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_220 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011382_010 |
0.0291481532 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 3 |
Hb_000010_160 |
0.0292139974 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 4 |
Hb_007187_010 |
0.0330806034 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 5 |
Hb_001519_140 |
0.0349771814 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 6 |
Hb_000731_150 |
0.0383411652 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 7 |
Hb_074312_010 |
0.041069627 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 8 |
Hb_014056_020 |
0.0416089166 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002965_170 |
0.0419173324 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 10 |
Hb_009707_020 |
0.0425544811 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 11 |
Hb_005610_070 |
0.0449364317 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 12 |
Hb_000512_120 |
0.0461705348 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 13 |
Hb_004993_020 |
0.0473328823 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_002328_080 |
0.0487825054 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_002743_010 |
0.0494210155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005280_020 |
0.0522274569 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 17 |
Hb_000844_080 |
0.0554968923 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 18 |
Hb_003086_140 |
0.0570307438 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 6-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_163566_020 |
0.0577696579 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000333_010 |
0.0601813622 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |