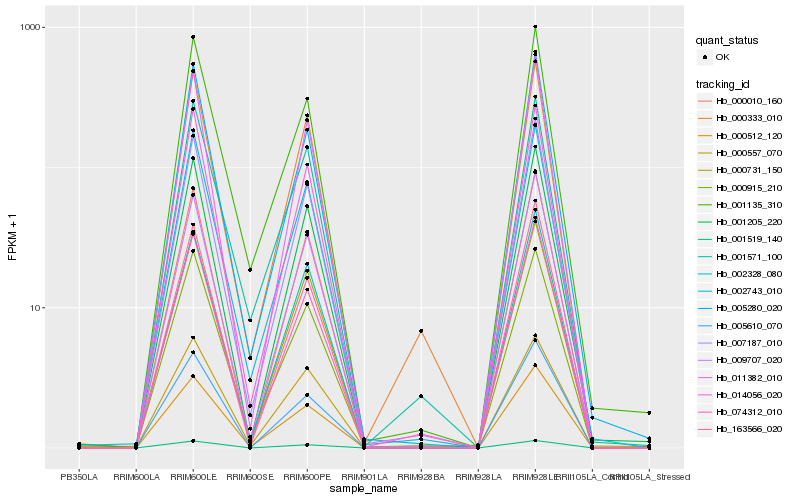
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000512_120 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 2 |
Hb_000333_010 |
0.0310463466 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 3 |
Hb_007187_010 |
0.0337692719 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 4 |
Hb_011382_010 |
0.0374599032 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 5 |
Hb_009707_020 |
0.0417438319 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 6 |
Hb_014056_020 |
0.0440211828 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002743_010 |
0.0449730014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001205_220 |
0.0461705348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000010_160 |
0.0472082712 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 10 |
Hb_002328_080 |
0.0479523169 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_074312_010 |
0.0487876353 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 12 |
Hb_000731_150 |
0.0505967336 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 13 |
Hb_005280_020 |
0.054439665 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 14 |
Hb_000915_210 |
0.0545410196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005610_070 |
0.0572306115 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 16 |
Hb_001571_100 |
0.057357143 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 17 |
Hb_001135_310 |
0.0578730515 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 18 |
Hb_163566_020 |
0.058548191 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_001519_140 |
0.0591992283 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 20 |
Hb_000557_070 |
0.061677625 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |