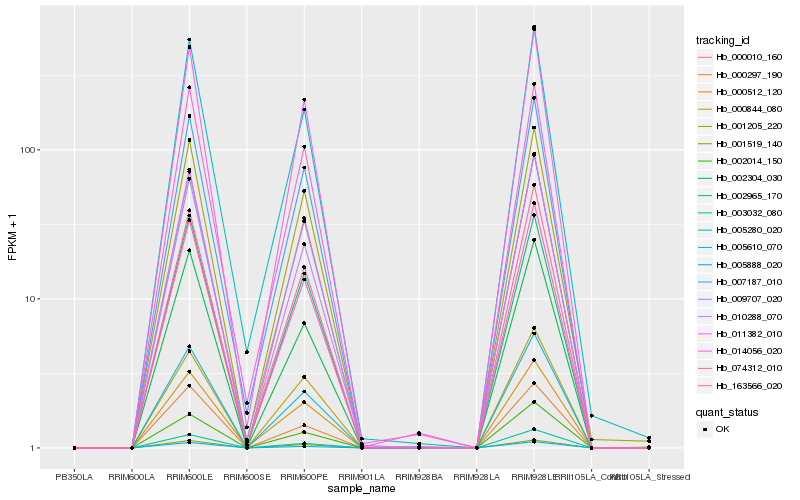
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074312_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 2 |
Hb_005610_070 |
0.016110222 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 3 |
Hb_011382_010 |
0.0251053021 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 4 |
Hb_163566_020 |
0.0253589767 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_007187_010 |
0.0278239833 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 6 |
Hb_002014_150 |
0.0308431154 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_010288_070 |
0.0326600419 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 8 |
Hb_005280_020 |
0.0353469598 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 9 |
Hb_000010_160 |
0.0377158592 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 10 |
Hb_002304_030 |
0.0390822944 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 11 |
Hb_003032_080 |
0.040380377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001205_220 |
0.041069627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009707_020 |
0.0429580586 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 14 |
Hb_000512_120 |
0.0487876353 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 15 |
Hb_005888_020 |
0.0501871033 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 16 |
Hb_014056_020 |
0.0534180256 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_002965_170 |
0.0576322038 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 18 |
Hb_000297_190 |
0.0582366108 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 19 |
Hb_001519_140 |
0.0583917017 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 20 |
Hb_000844_080 |
0.0604269229 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |