| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
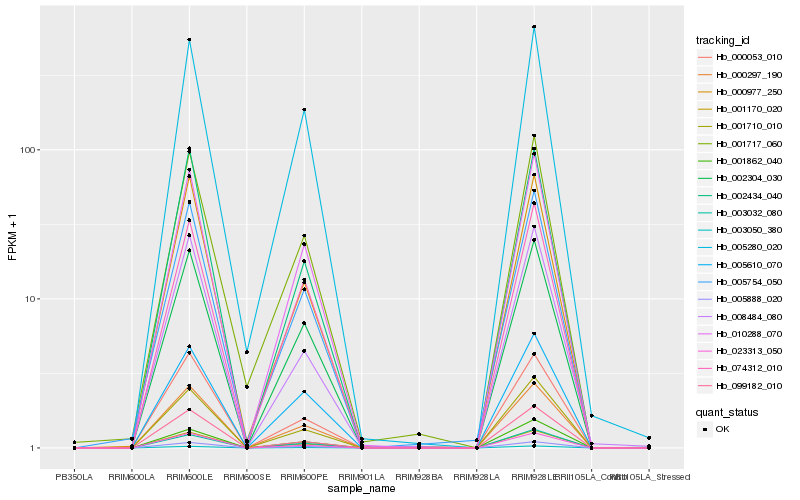
Hb_003050_380 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 2 |
Hb_001170_020 |
0.0205918401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005888_020 |
0.0279019552 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 4 |
Hb_010288_070 |
0.0363725524 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 5 |
Hb_002304_030 |
0.0374330298 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 6 |
Hb_000297_190 |
0.0417142145 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 7 |
Hb_003032_080 |
0.042262709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005754_050 |
0.0470643523 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 9 |
Hb_000977_250 |
0.0514061735 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_023313_050 |
0.0516488511 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 11 |
Hb_001862_040 |
0.0528923387 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 12 |
Hb_002434_040 |
0.0543200451 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 13 |
Hb_000053_010 |
0.0560527413 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 14 |
Hb_001710_010 |
0.0581901498 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 15 |
Hb_005610_070 |
0.0588998369 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 16 |
Hb_005280_020 |
0.0651186311 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 17 |
Hb_074312_010 |
0.0652583642 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 18 |
Hb_099182_010 |
0.065577811 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 19 |
Hb_008484_080 |
0.0674421083 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001717_060 |
0.0677854967 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |