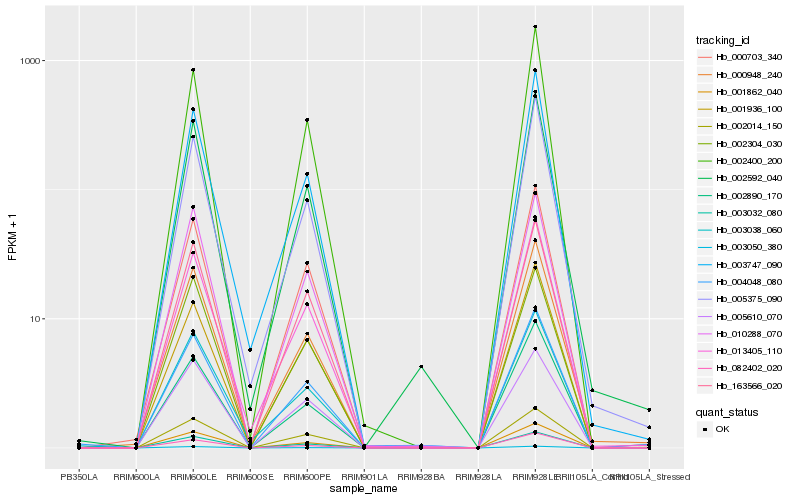
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_040 |
0.0 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 2 |
Hb_003032_080 |
0.031885899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_082402_020 |
0.0403006926 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 4 |
Hb_000948_240 |
0.0500794008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002890_170 |
0.0513194188 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 6 |
Hb_002014_150 |
0.0514142384 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_003050_380 |
0.0528923387 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 8 |
Hb_163566_020 |
0.0547391745 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_010288_070 |
0.0553909583 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 10 |
Hb_002400_200 |
0.0563694593 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 11 |
Hb_000703_340 |
0.0583989064 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_002592_040 |
0.0596544229 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 13 |
Hb_013405_110 |
0.0599336083 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 14 |
Hb_003747_090 |
0.0599767305 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_004048_080 |
0.0609306579 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 16 |
Hb_001936_100 |
0.0635588604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005375_090 |
0.0641508376 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003038_060 |
0.0654800317 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 19 |
Hb_005610_070 |
0.0690815767 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 20 |
Hb_002304_030 |
0.0694871715 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |