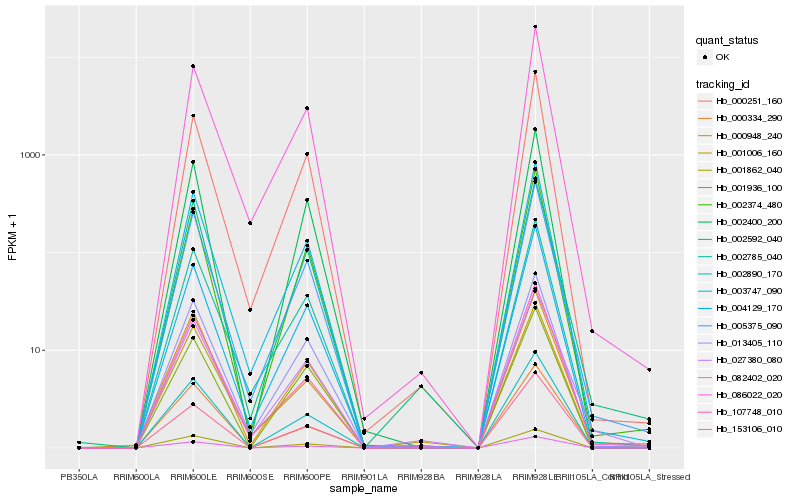
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005375_090 |
0.0 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003747_090 |
0.0197979599 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_002785_040 |
0.0356770946 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_013405_110 |
0.0386114486 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_002890_170 |
0.0475502087 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 6 |
Hb_004129_170 |
0.0479635893 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 7 |
Hb_082402_020 |
0.048827541 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 8 |
Hb_086022_020 |
0.0492738839 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_002374_480 |
0.0517390951 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_002400_200 |
0.0525348568 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 11 |
Hb_000948_240 |
0.0528024846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002592_040 |
0.0584871931 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 13 |
Hb_027380_080 |
0.0596719729 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 14 |
Hb_001006_160 |
0.0615984342 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_000251_160 |
0.0618373726 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 16 |
Hb_107748_010 |
0.0621602186 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001862_040 |
0.0641508376 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 18 |
Hb_001936_100 |
0.0684858337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000334_290 |
0.069216933 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 20 |
Hb_153106_010 |
0.0697655378 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |