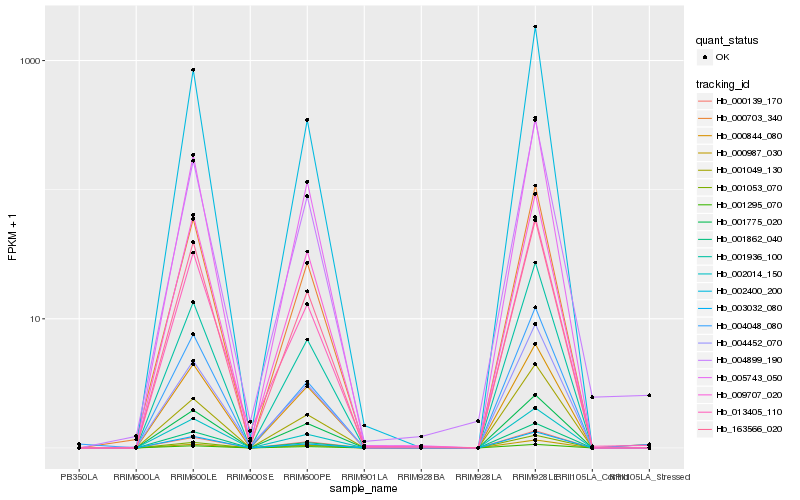
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_340 |
0.0 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001936_100 |
0.0385429732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002014_150 |
0.0465155125 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 4 |
Hb_163566_020 |
0.0486814689 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002400_200 |
0.0488301123 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 6 |
Hb_000139_170 |
0.0496669691 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 7 |
Hb_005743_050 |
0.0515824892 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_013405_110 |
0.0538493928 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 9 |
Hb_004452_070 |
0.0560227415 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_003032_080 |
0.0580439594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001862_040 |
0.0583989064 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 12 |
Hb_001775_020 |
0.0592869415 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 13 |
Hb_001053_070 |
0.0594422127 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein EUGRSUZ_B02976 [Eucalyptus grandis] |
| 14 |
Hb_004899_190 |
0.0598723138 |
- |
- |
RecName: Full=Peroxiredoxin Q, chloroplastic; AltName: Full=Thioredoxin reductase; Flags: Precursor [Populus x jackii] |
| 15 |
Hb_001049_130 |
0.060609385 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000987_030 |
0.0654942145 |
transcription factor |
TF Family: BES1 |
hypothetical protein POPTR_0016s13350g [Populus trichocarpa] |
| 17 |
Hb_004048_080 |
0.0665530453 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 18 |
Hb_009707_020 |
0.0669801468 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 19 |
Hb_000844_080 |
0.0686277006 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 20 |
Hb_001295_070 |
0.0688581665 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |