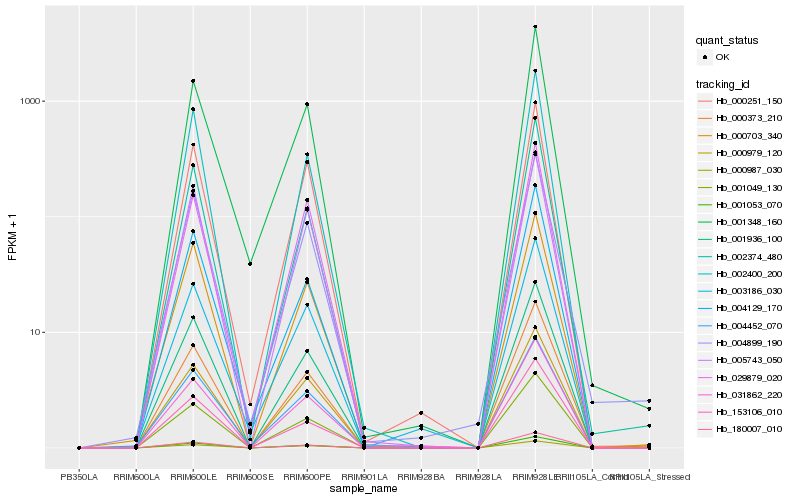
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001053_070 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein EUGRSUZ_B02976 [Eucalyptus grandis] |
| 2 |
Hb_001049_130 |
0.0060015576 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_031862_220 |
0.0169338546 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 4 |
Hb_000373_210 |
0.0247130059 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 5 |
Hb_001936_100 |
0.0272032313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002400_200 |
0.0391628259 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 7 |
Hb_000987_030 |
0.0463046002 |
transcription factor |
TF Family: BES1 |
hypothetical protein POPTR_0016s13350g [Populus trichocarpa] |
| 8 |
Hb_002374_480 |
0.0498596111 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000251_150 |
0.052299453 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_180007_010 |
0.0547507114 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_004452_070 |
0.0560818604 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_005743_050 |
0.0589741347 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_000703_340 |
0.0594422127 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000979_120 |
0.0618840228 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_153106_010 |
0.0641537699 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 16 |
Hb_004129_170 |
0.0647127756 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 17 |
Hb_001348_160 |
0.065089906 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_003186_030 |
0.0671785527 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0012s05720g [Populus trichocarpa] |
| 19 |
Hb_029879_020 |
0.0693691199 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 20 |
Hb_004899_190 |
0.0701645811 |
- |
- |
RecName: Full=Peroxiredoxin Q, chloroplastic; AltName: Full=Thioredoxin reductase; Flags: Precursor [Populus x jackii] |