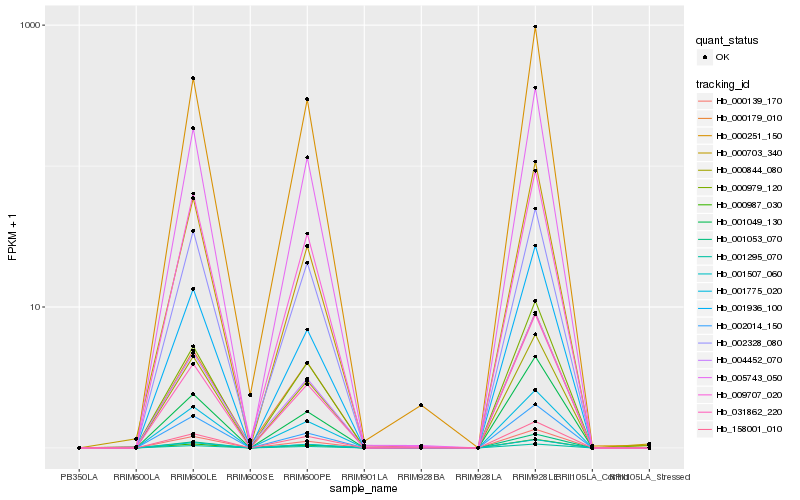
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005743_050 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_000139_170 |
0.0241488274 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 3 |
Hb_000987_030 |
0.027612739 |
transcription factor |
TF Family: BES1 |
hypothetical protein POPTR_0016s13350g [Populus trichocarpa] |
| 4 |
Hb_001775_020 |
0.0333222866 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 5 |
Hb_001295_070 |
0.0337257462 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 6 |
Hb_158001_010 |
0.0385813311 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 7 |
Hb_000251_150 |
0.0398368305 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000844_080 |
0.0435437442 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 9 |
Hb_001507_060 |
0.0503747367 |
- |
- |
translation initiation factor eif-2b gamma subunit, putative [Ricinus communis] |
| 10 |
Hb_001936_100 |
0.0515121189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000703_340 |
0.0515824892 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000179_010 |
0.0551346991 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 13 |
Hb_001049_130 |
0.0555284107 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000979_120 |
0.0558907401 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_009707_020 |
0.0566766723 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 16 |
Hb_004452_070 |
0.0588928202 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001053_070 |
0.0589741347 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein EUGRSUZ_B02976 [Eucalyptus grandis] |
| 18 |
Hb_002328_080 |
0.0619061531 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_002014_150 |
0.0643419142 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 20 |
Hb_031862_220 |
0.0654293407 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |