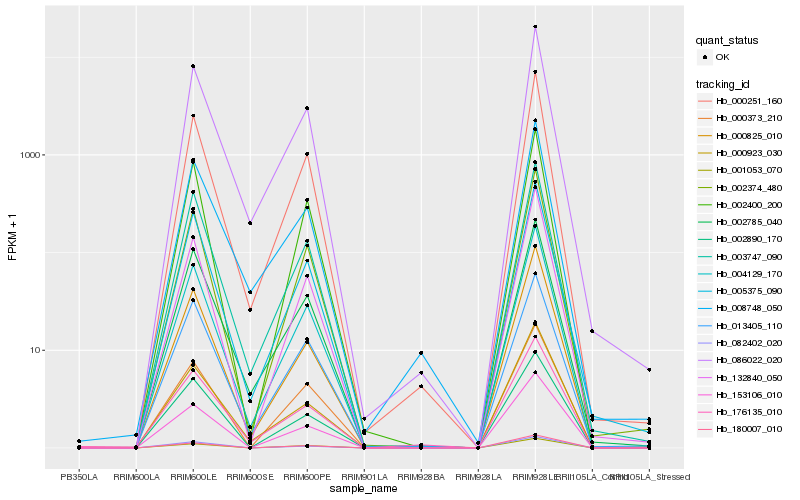
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_170 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 2 |
Hb_000251_160 |
0.0248138801 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 3 |
Hb_002374_480 |
0.0330828897 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_086022_020 |
0.0338539655 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_153106_010 |
0.0373541998 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 6 |
Hb_180007_010 |
0.0428276381 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_003747_090 |
0.045802573 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_005375_090 |
0.0479635893 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002400_200 |
0.0484163384 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 10 |
Hb_002890_170 |
0.0504641133 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 11 |
Hb_000825_010 |
0.0540366116 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 12 |
Hb_002785_040 |
0.0542139294 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 13 |
Hb_000373_210 |
0.057236008 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 14 |
Hb_132840_050 |
0.0588931449 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 15 |
Hb_082402_020 |
0.060275341 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 16 |
Hb_013405_110 |
0.0628597968 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_001053_070 |
0.0647127756 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein EUGRSUZ_B02976 [Eucalyptus grandis] |
| 18 |
Hb_008748_050 |
0.0652567849 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000923_030 |
0.0657252639 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 20 |
Hb_176135_010 |
0.0666049833 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |