| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
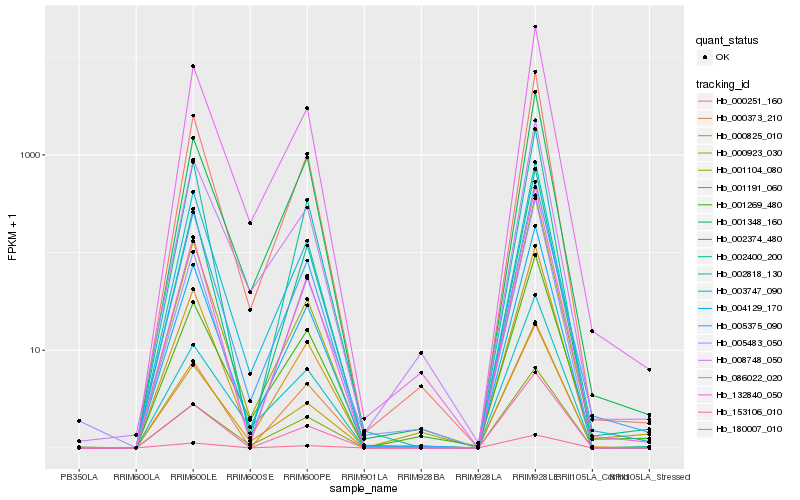
Hb_000251_160 |
0.0 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 2 |
Hb_004129_170 |
0.0248138801 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 3 |
Hb_086022_020 |
0.0331451407 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 4 |
Hb_002374_480 |
0.0358748313 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_153106_010 |
0.0382826279 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 6 |
Hb_180007_010 |
0.0394295487 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_132840_050 |
0.0451927806 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000825_010 |
0.0481197514 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 9 |
Hb_000923_030 |
0.0531163154 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 10 |
Hb_000373_210 |
0.0596560103 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 11 |
Hb_001269_480 |
0.0601842187 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001348_160 |
0.0602420414 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_005483_050 |
0.0608199623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008748_050 |
0.0611631053 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_005375_090 |
0.0618373726 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003747_090 |
0.0628370407 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_001104_080 |
0.0630594139 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 18 |
Hb_002400_200 |
0.0644550571 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 19 |
Hb_001191_060 |
0.064483046 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 20 |
Hb_002818_130 |
0.0658599508 |
- |
- |
cytochrome P450, putative [Ricinus communis] |