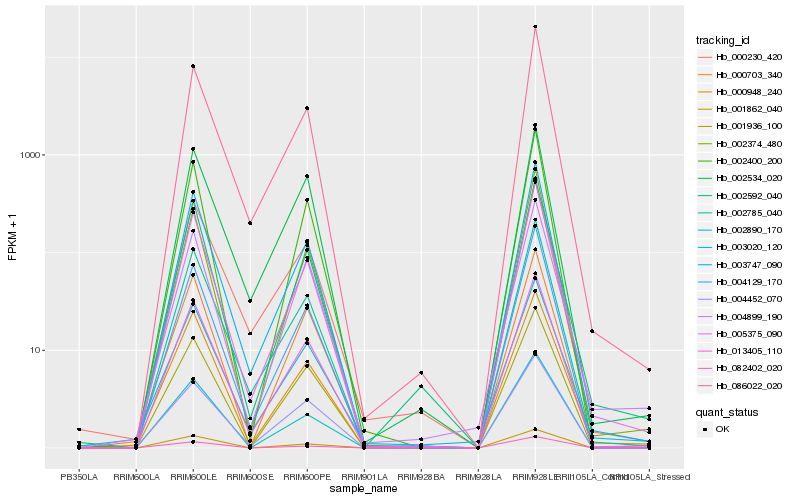
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013405_110 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_003747_090 |
0.0340094174 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_002785_040 |
0.0364792839 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_005375_090 |
0.0386114486 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002400_200 |
0.0506294734 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 6 |
Hb_000703_340 |
0.0538493928 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002592_040 |
0.0550204062 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 8 |
Hb_001936_100 |
0.0564415629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000948_240 |
0.0593204074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001862_040 |
0.0599336083 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 11 |
Hb_004452_070 |
0.0612967971 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_003020_120 |
0.0619307054 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 13 |
Hb_004129_170 |
0.0628597968 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 14 |
Hb_086022_020 |
0.063596684 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 15 |
Hb_002534_020 |
0.0636646957 |
- |
- |
PREDICTED: chlorophyll a-b binding protein 4, chloroplastic [Gossypium raimondii] |
| 16 |
Hb_000230_420 |
0.0641660676 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 17 |
Hb_082402_020 |
0.0641757739 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 18 |
Hb_004899_190 |
0.0653500149 |
- |
- |
RecName: Full=Peroxiredoxin Q, chloroplastic; AltName: Full=Thioredoxin reductase; Flags: Precursor [Populus x jackii] |
| 19 |
Hb_002374_480 |
0.0656708386 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_002890_170 |
0.065837797 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |