| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
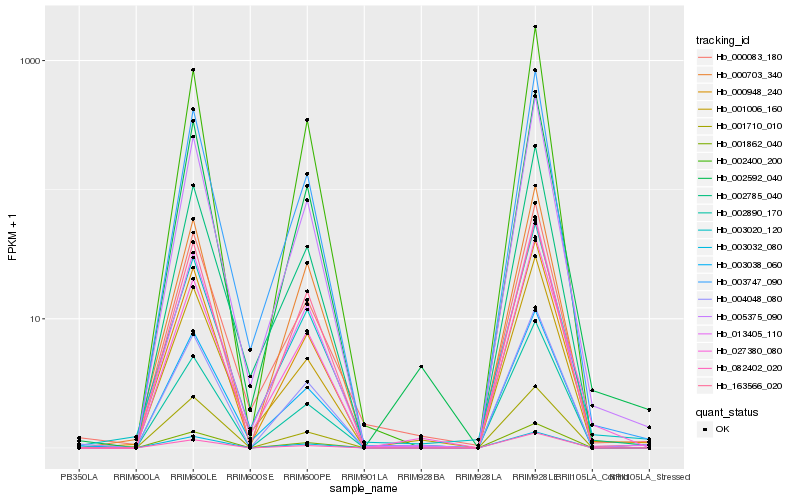
Hb_002592_040 |
0.0 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 2 |
Hb_000948_240 |
0.0484737396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_013405_110 |
0.0550204062 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 4 |
Hb_005375_090 |
0.0584871931 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001006_160 |
0.0596223991 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001862_040 |
0.0596544229 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 7 |
Hb_003747_090 |
0.0631924569 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_003032_080 |
0.0681892763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_027380_080 |
0.0684698288 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 10 |
Hb_003020_120 |
0.0697398672 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 11 |
Hb_082402_020 |
0.0705822278 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 12 |
Hb_004048_080 |
0.0706219368 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 13 |
Hb_001710_010 |
0.0709374419 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 14 |
Hb_000083_180 |
0.0711059776 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 15 |
Hb_002785_040 |
0.0715828814 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 16 |
Hb_000703_340 |
0.0728679139 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_003038_060 |
0.0749169051 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 18 |
Hb_163566_020 |
0.075187555 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_002400_200 |
0.076105619 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 20 |
Hb_002890_170 |
0.076357515 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |