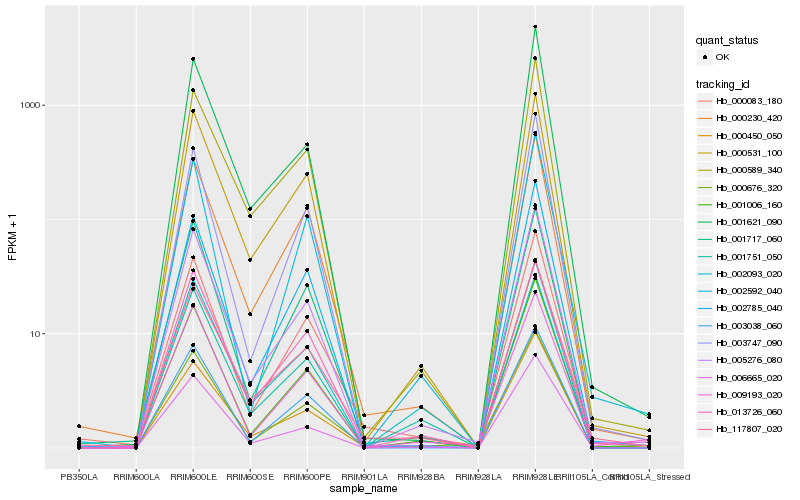
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005276_080 |
0.0 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001006_160 |
0.0363835137 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_006665_020 |
0.0500516252 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 4 |
Hb_000531_100 |
0.0552563451 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000450_050 |
0.058748385 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 6 |
Hb_000589_340 |
0.0618022899 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 7 |
Hb_000676_320 |
0.0623088798 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 8 |
Hb_000083_180 |
0.0623455224 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 9 |
Hb_117807_020 |
0.0624858822 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_009193_020 |
0.0635132001 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 11 |
Hb_001621_090 |
0.0642967586 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 12 |
Hb_002785_040 |
0.0658685791 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 13 |
Hb_003038_060 |
0.068248969 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 14 |
Hb_000230_420 |
0.0705703976 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 15 |
Hb_001751_050 |
0.0725096099 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003747_090 |
0.073800964 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_001717_060 |
0.0739753182 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 18 |
Hb_013726_060 |
0.0752955729 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 19 |
Hb_002093_020 |
0.0761997684 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 20 |
Hb_002592_040 |
0.0765700644 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |