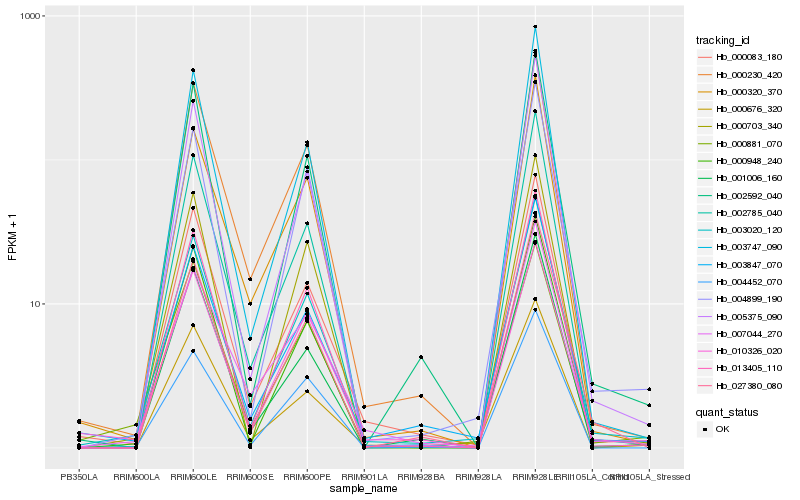
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_120 |
0.0 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 2 |
Hb_000083_180 |
0.059759866 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 3 |
Hb_013405_110 |
0.0619307054 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 4 |
Hb_004899_190 |
0.0630341636 |
- |
- |
RecName: Full=Peroxiredoxin Q, chloroplastic; AltName: Full=Thioredoxin reductase; Flags: Precursor [Populus x jackii] |
| 5 |
Hb_000948_240 |
0.0686432556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003847_070 |
0.0696576508 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002592_040 |
0.0697398672 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 8 |
Hb_005375_090 |
0.0707443237 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002785_040 |
0.0716239387 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 10 |
Hb_000230_420 |
0.0730966721 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 11 |
Hb_003747_090 |
0.0750290345 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_027380_080 |
0.077052196 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 13 |
Hb_000703_340 |
0.079658768 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001006_160 |
0.0805419866 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_010326_020 |
0.0812652872 |
- |
- |
PREDICTED: thioredoxin-like 4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007044_270 |
0.081916891 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000881_070 |
0.0823367654 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000676_320 |
0.0827861962 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 19 |
Hb_004452_070 |
0.0832200733 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000320_370 |
0.0841197 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |