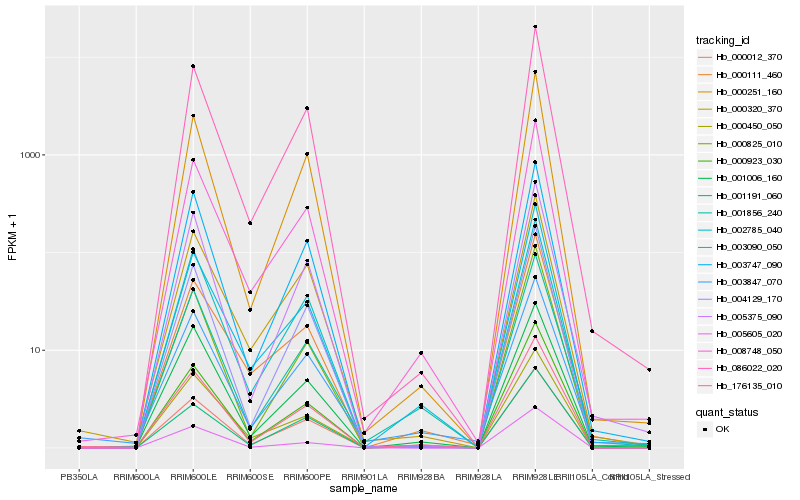
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176135_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_008748_050 |
0.0434799635 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_000012_370 |
0.0536506564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_086022_020 |
0.0580341663 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_003847_070 |
0.0601501924 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002785_040 |
0.0657480468 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 7 |
Hb_000251_160 |
0.0662765682 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 8 |
Hb_001856_240 |
0.0665083644 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_004129_170 |
0.0666049833 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 10 |
Hb_000320_370 |
0.067487714 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003090_050 |
0.0690915869 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 12 |
Hb_000923_030 |
0.069472073 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 13 |
Hb_003747_090 |
0.0719758483 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_001006_160 |
0.0738708652 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_000825_010 |
0.0761192053 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 16 |
Hb_005375_090 |
0.0769210769 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000450_050 |
0.0772912782 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 18 |
Hb_001191_060 |
0.0774830088 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 19 |
Hb_005605_020 |
0.0777999693 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_000111_460 |
0.078187723 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |