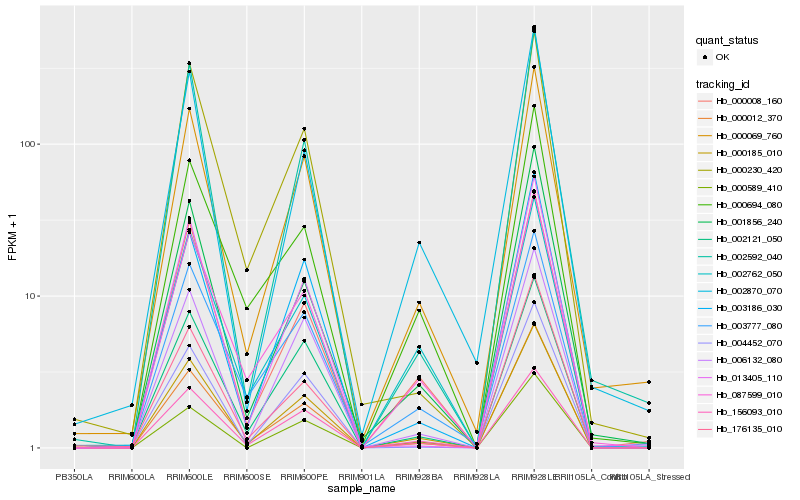
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000185_010 |
0.0 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 2 |
Hb_000069_760 |
0.0625437631 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000012_370 |
0.066769226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000008_160 |
0.0691063161 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 5 |
Hb_087599_010 |
0.0699471418 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 6 |
Hb_156093_010 |
0.0719813122 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 7 |
Hb_006132_080 |
0.0790527012 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 8 |
Hb_002121_050 |
0.0804623915 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |
| 9 |
Hb_000589_410 |
0.084025001 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic-like isoform X1 [Populus euphratica] |
| 10 |
Hb_003186_030 |
0.0842728481 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0012s05720g [Populus trichocarpa] |
| 11 |
Hb_002870_070 |
0.0851503634 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 12 |
Hb_003777_080 |
0.085844218 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 13 |
Hb_002762_050 |
0.0892929761 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 14 |
Hb_002592_040 |
0.0896483643 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 15 |
Hb_176135_010 |
0.0898433911 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_001856_240 |
0.0918556386 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 17 |
Hb_000230_420 |
0.0922934427 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 18 |
Hb_013405_110 |
0.0934849809 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 19 |
Hb_000694_080 |
0.0936364934 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 20 |
Hb_004452_070 |
0.0943639081 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |