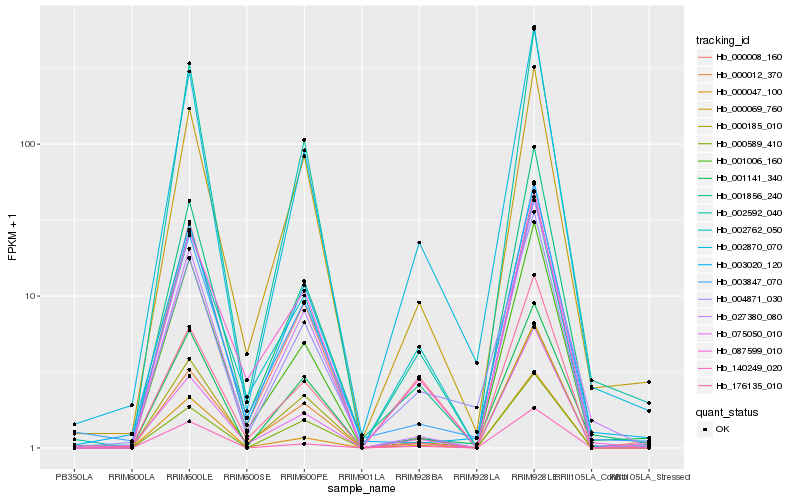
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002870_070 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_004871_030 |
0.0667396747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001856_240 |
0.072435848 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 4 |
Hb_000008_160 |
0.0747214528 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 5 |
Hb_000185_010 |
0.0851503634 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 6 |
Hb_000069_760 |
0.0884772652 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003847_070 |
0.0898088733 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002592_040 |
0.0929498381 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 9 |
Hb_001006_160 |
0.1003449881 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_000047_100 |
0.1021333438 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_140249_020 |
0.102517065 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 12 |
Hb_000012_370 |
0.1035929243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000589_410 |
0.1051436526 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic-like isoform X1 [Populus euphratica] |
| 14 |
Hb_027380_080 |
0.1059508549 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 15 |
Hb_176135_010 |
0.1066086974 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_075050_010 |
0.107055773 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003020_120 |
0.1084384077 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 18 |
Hb_001141_340 |
0.1091546648 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 19 |
Hb_002762_050 |
0.1091952766 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 20 |
Hb_087599_010 |
0.1116951665 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |