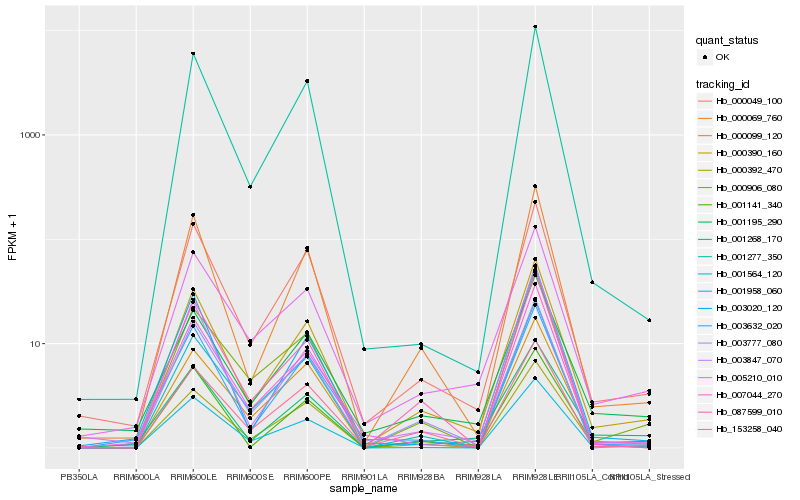
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_160 |
0.0 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 2 |
Hb_000069_760 |
0.0608600239 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000049_100 |
0.0692077222 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 4 |
Hb_003777_080 |
0.0824379759 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 5 |
Hb_001141_340 |
0.0858662477 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 6 |
Hb_001958_060 |
0.0866461432 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_005210_010 |
0.0895850534 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 8 |
Hb_153258_040 |
0.091009618 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 9 |
Hb_001268_170 |
0.0922027967 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 10 |
Hb_003020_120 |
0.0944548906 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 11 |
Hb_000099_120 |
0.0950643267 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 12 |
Hb_000392_470 |
0.0950861905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007044_270 |
0.0950933145 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000906_080 |
0.0954378317 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_001564_120 |
0.0964328588 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 16 |
Hb_087599_010 |
0.0983150775 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 17 |
Hb_003632_020 |
0.099205477 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 18 |
Hb_001277_350 |
0.0993181938 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 19 |
Hb_001195_290 |
0.0996283895 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 20 |
Hb_003847_070 |
0.10002313 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |