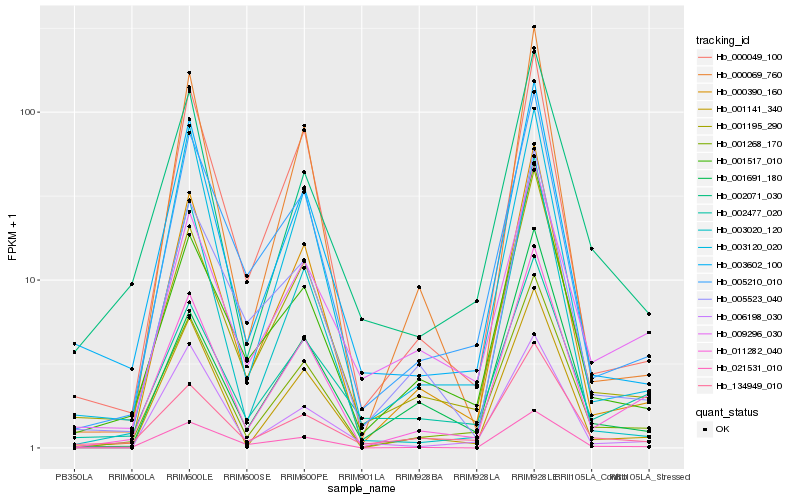
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_170 |
0.0 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 2 |
Hb_011282_040 |
0.0740437916 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 3 |
Hb_000390_160 |
0.0922027967 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 4 |
Hb_001195_290 |
0.0924465316 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 5 |
Hb_001141_340 |
0.102666677 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 6 |
Hb_005210_010 |
0.1071007345 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 7 |
Hb_002071_030 |
0.1153470938 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 8 |
Hb_003602_100 |
0.1179896419 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_001517_010 |
0.1181246834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_134949_010 |
0.1215554172 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 11 |
Hb_000049_100 |
0.1235058212 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 12 |
Hb_003120_020 |
0.1241258768 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 13 |
Hb_000069_760 |
0.1305701966 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003020_120 |
0.1335740115 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 15 |
Hb_021531_010 |
0.1366018504 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_009296_030 |
0.136675133 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 17 |
Hb_006198_030 |
0.1371221512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001691_180 |
0.1373154829 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 19 |
Hb_005523_040 |
0.1375154139 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 20 |
Hb_002477_020 |
0.1375646256 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |