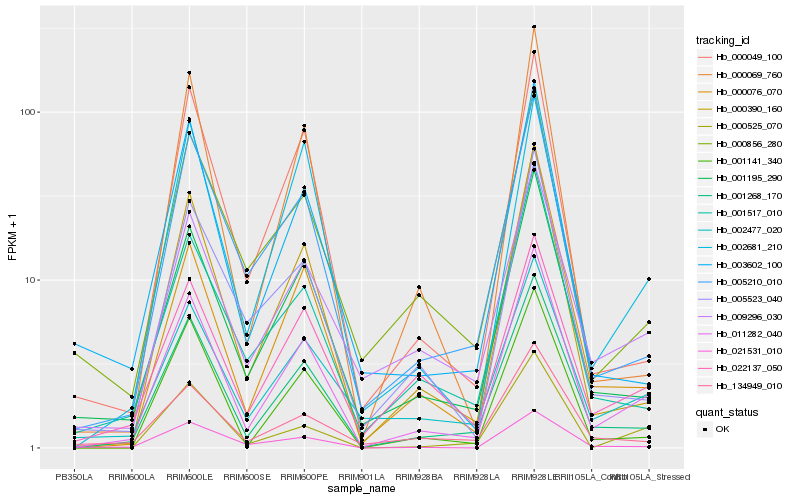
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011282_040 |
0.0 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 2 |
Hb_001268_170 |
0.0740437916 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 3 |
Hb_000390_160 |
0.1046969328 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 4 |
Hb_001195_290 |
0.1080416498 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 5 |
Hb_001141_340 |
0.1202693195 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 6 |
Hb_002477_020 |
0.1261641687 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 7 |
Hb_005210_010 |
0.1262190145 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 8 |
Hb_022137_050 |
0.1283713005 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 9 |
Hb_009296_030 |
0.130203064 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 10 |
Hb_001517_010 |
0.1333792875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000049_100 |
0.1355278223 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 12 |
Hb_002681_210 |
0.1360777906 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 13 |
Hb_021531_010 |
0.1382894374 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 14 |
Hb_005523_040 |
0.1397578351 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 15 |
Hb_000525_070 |
0.1399438978 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 16 |
Hb_000069_760 |
0.1404483058 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003602_100 |
0.141295115 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000076_070 |
0.1441723687 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 19 |
Hb_134949_010 |
0.1457938115 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 20 |
Hb_000856_280 |
0.1469598604 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |