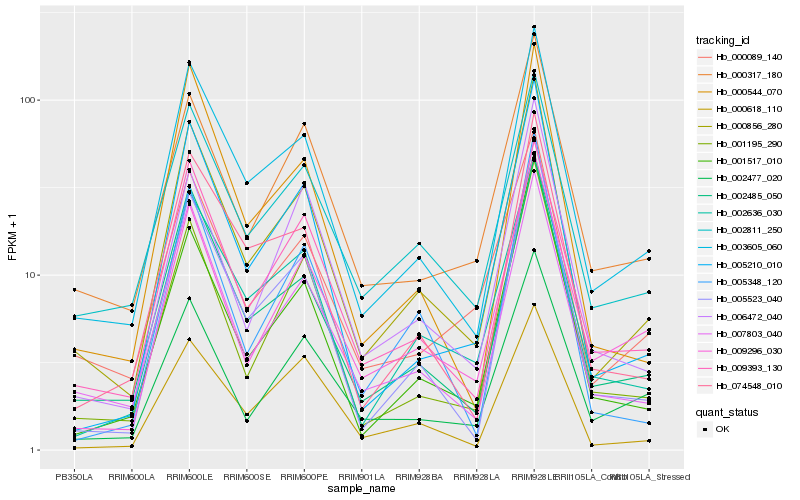
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 2 |
Hb_003605_060 |
0.0683222318 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002485_050 |
0.0760068061 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007803_040 |
0.0780086395 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005523_040 |
0.0906683715 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 6 |
Hb_005210_010 |
0.0914598059 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 7 |
Hb_001195_290 |
0.0938129681 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 8 |
Hb_002636_030 |
0.0944158127 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 9 |
Hb_009393_130 |
0.1017522603 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 10 |
Hb_000089_140 |
0.1022743695 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 11 |
Hb_074548_010 |
0.1034358795 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_001517_010 |
0.1034903339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002811_250 |
0.105826717 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000544_070 |
0.1072588293 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 15 |
Hb_009296_030 |
0.1089480834 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 16 |
Hb_000317_180 |
0.1090732172 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000618_110 |
0.1117103888 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 18 |
Hb_005348_120 |
0.1126365732 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006472_040 |
0.1131657906 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002477_020 |
0.1140459052 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |