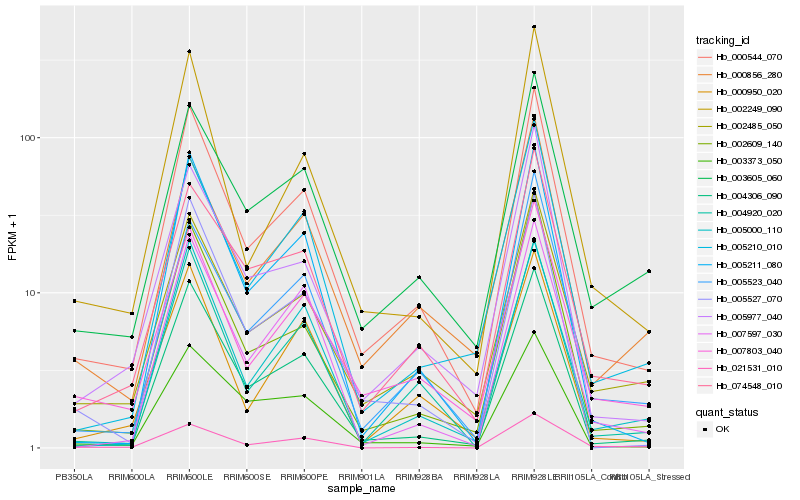
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000544_070 |
0.0 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 2 |
Hb_004306_090 |
0.0723823176 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002485_050 |
0.0821892263 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002609_140 |
0.0839519575 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_007803_040 |
0.0873276279 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004920_020 |
0.0905718664 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 7 |
Hb_074548_010 |
0.0939470829 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_005527_070 |
0.0955756952 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 9 |
Hb_002249_090 |
0.0956036629 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005977_040 |
0.0974495075 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 11 |
Hb_005523_040 |
0.0988126532 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 12 |
Hb_005211_080 |
0.0995263524 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 13 |
Hb_003605_060 |
0.102059795 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000950_020 |
0.1026144646 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 15 |
Hb_021531_010 |
0.1059774495 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_005210_010 |
0.1068115675 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 17 |
Hb_000856_280 |
0.1072588293 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 18 |
Hb_005000_110 |
0.1080500029 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007597_030 |
0.1088259425 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003373_050 |
0.1088911228 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |