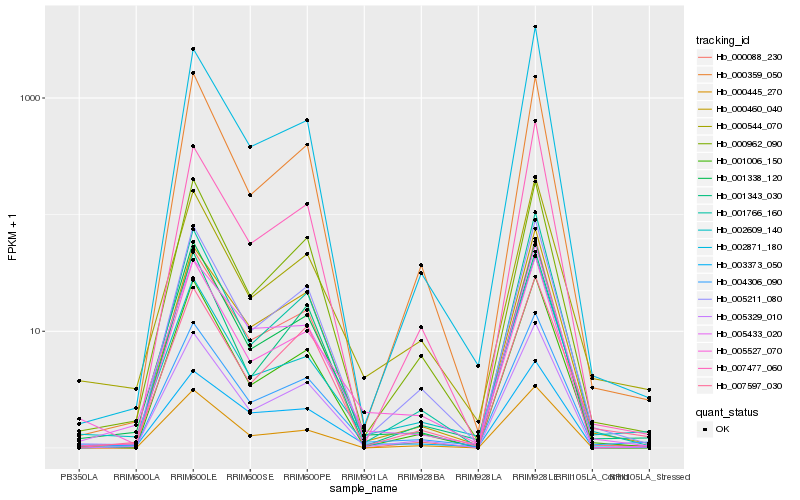
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004306_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005211_080 |
0.0638070897 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 3 |
Hb_005329_010 |
0.066583253 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 4 |
Hb_000544_070 |
0.0723823176 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 5 |
Hb_000460_040 |
0.0764782168 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 6 |
Hb_001006_150 |
0.0788287818 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_005433_020 |
0.0816282274 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 8 |
Hb_005527_070 |
0.0816937972 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 9 |
Hb_001338_120 |
0.0826656966 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 10 |
Hb_001766_160 |
0.0830820349 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 11 |
Hb_000962_090 |
0.0836975346 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 12 |
Hb_007597_030 |
0.0865999719 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002609_140 |
0.0876817986 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000088_230 |
0.0886925758 |
- |
- |
hypothetical protein POPTR_0013s11340g [Populus trichocarpa] |
| 15 |
Hb_001343_030 |
0.0898361383 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002871_180 |
0.0906497542 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000359_050 |
0.0924193947 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_003373_050 |
0.0928454149 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 19 |
Hb_007477_060 |
0.0952498353 |
- |
- |
unknown [Medicago truncatula] |
| 20 |
Hb_000445_270 |
0.0958655681 |
- |
- |
ATP binding protein, putative [Ricinus communis] |