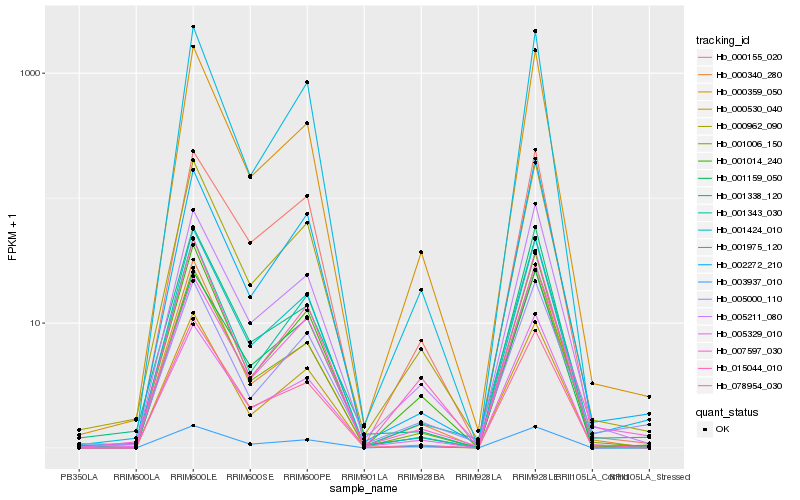
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000962_090 |
0.0 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 2 |
Hb_005211_080 |
0.0443611449 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 3 |
Hb_000359_050 |
0.0458143798 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001343_030 |
0.052305627 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001014_240 |
0.0567644589 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 6 |
Hb_005329_010 |
0.0698126553 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 7 |
Hb_001006_150 |
0.0703314052 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001424_010 |
0.0709271945 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001975_120 |
0.0718862818 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 10 |
Hb_001159_050 |
0.0722099783 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 11 |
Hb_000340_280 |
0.0773441901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000530_040 |
0.0781018919 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001338_120 |
0.0791158053 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 14 |
Hb_078954_030 |
0.0796372233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005000_110 |
0.0798920969 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000155_020 |
0.0814974859 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_007597_030 |
0.0816870591 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003937_010 |
0.0817974641 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 19 |
Hb_015044_010 |
0.0822870461 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 20 |
Hb_002272_210 |
0.083301038 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |