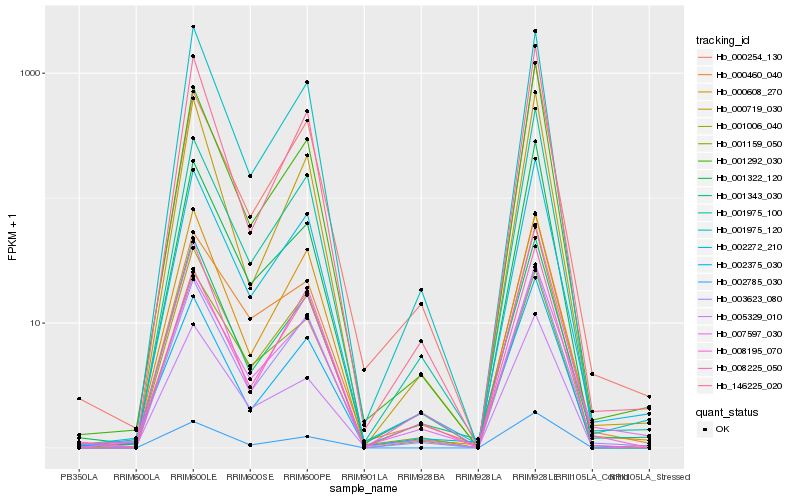
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002272_210 |
0.0 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003623_080 |
0.03691464 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 3 |
Hb_001343_030 |
0.0588057735 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007597_030 |
0.0612843409 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001006_040 |
0.0613173504 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 6 |
Hb_146225_020 |
0.0659314952 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 7 |
Hb_001975_120 |
0.069132187 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 8 |
Hb_001159_050 |
0.0692600023 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 9 |
Hb_001292_030 |
0.0704631883 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000254_130 |
0.0724524279 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001975_100 |
0.0732275632 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008225_050 |
0.07438284 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 13 |
Hb_000608_270 |
0.0747602838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002375_030 |
0.0762812554 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 15 |
Hb_000460_040 |
0.0762932458 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 16 |
Hb_002785_030 |
0.0763590086 |
- |
- |
Polyketide cyclase/dehydrase and lipid transport superfamily protein [Theobroma cacao] |
| 17 |
Hb_000719_030 |
0.0771208698 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_008195_070 |
0.0788580366 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_005329_010 |
0.0794929032 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 20 |
Hb_001322_120 |
0.0809769821 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |