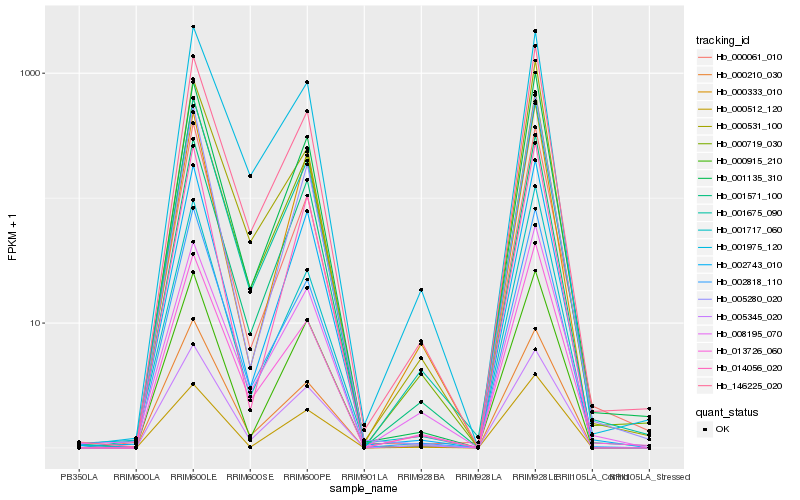
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000719_030 |
0.0 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_146225_020 |
0.0206804578 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 3 |
Hb_001135_310 |
0.031228419 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 4 |
Hb_001675_090 |
0.0367990198 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_002818_110 |
0.0487733551 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 6 |
Hb_013726_060 |
0.0504558256 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 7 |
Hb_001571_100 |
0.0512436505 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 8 |
Hb_002743_010 |
0.0543606764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005345_020 |
0.0548864528 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 10 |
Hb_005280_020 |
0.0601429623 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 11 |
Hb_000061_010 |
0.0622692714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000915_210 |
0.0633126947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000512_120 |
0.0656281971 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 14 |
Hb_001975_120 |
0.0667858358 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 15 |
Hb_014056_020 |
0.0672288164 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_008195_070 |
0.0672676969 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_001717_060 |
0.068310467 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 18 |
Hb_000210_030 |
0.0699376464 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 19 |
Hb_000531_100 |
0.0706108072 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000333_010 |
0.0718602411 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |