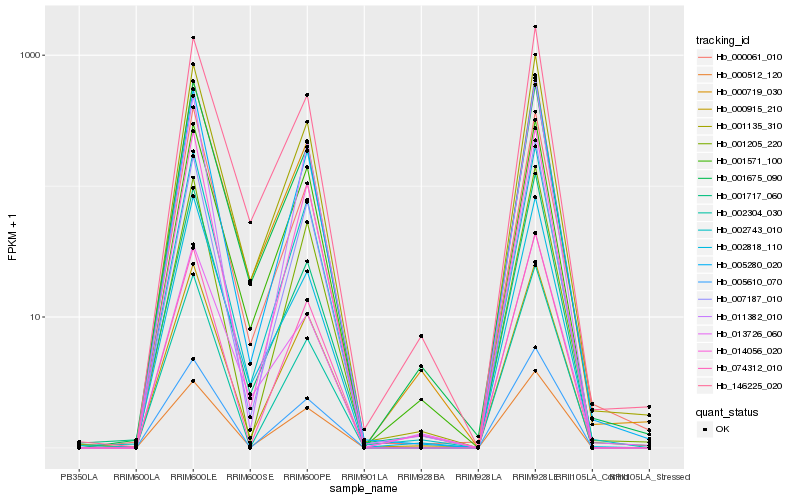
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001135_310 |
0.0 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 2 |
Hb_000719_030 |
0.031228419 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_146225_020 |
0.035765765 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 4 |
Hb_005280_020 |
0.0380413951 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 5 |
Hb_002743_010 |
0.0431238527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002818_110 |
0.0549517683 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 7 |
Hb_000915_210 |
0.0568536452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_014056_020 |
0.0568948833 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_007187_010 |
0.0569270685 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 10 |
Hb_001675_090 |
0.0571470656 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000512_120 |
0.0578730515 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 12 |
Hb_001571_100 |
0.0579946965 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 13 |
Hb_000061_010 |
0.0599918768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001717_060 |
0.0624627478 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 15 |
Hb_074312_010 |
0.062639654 |
- |
- |
PREDICTED: cytochrome P450 82A3-like [Jatropha curcas] |
| 16 |
Hb_013726_060 |
0.0639598536 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 17 |
Hb_001205_220 |
0.065722223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011382_010 |
0.0675696684 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 19 |
Hb_005610_070 |
0.0692077639 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 20 |
Hb_002304_030 |
0.0706821425 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |