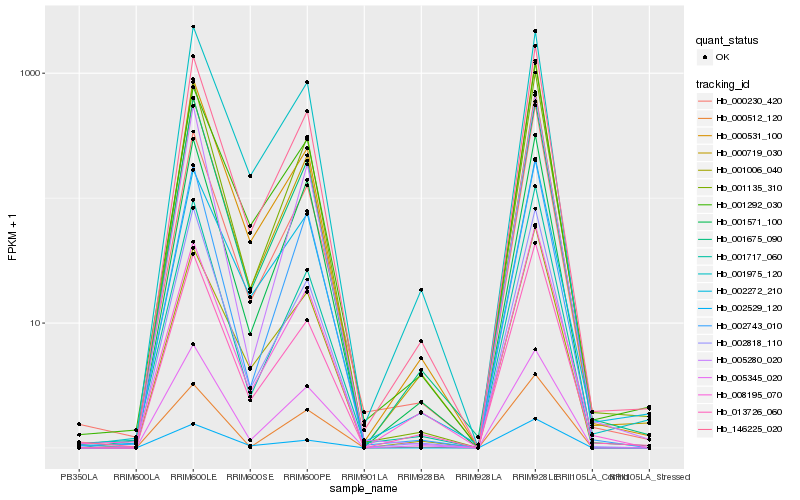
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146225_020 |
0.0 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 2 |
Hb_000719_030 |
0.0206804578 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001135_310 |
0.035765765 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 4 |
Hb_013726_060 |
0.045658585 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 5 |
Hb_001675_090 |
0.0531730873 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000531_100 |
0.0583781641 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_001571_100 |
0.058479011 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 8 |
Hb_002818_110 |
0.0589980565 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 9 |
Hb_008195_070 |
0.0591198411 |
- |
- |
PREDICTED: thylakoid lumenal protein At1g12250, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_001006_040 |
0.0610493161 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 11 |
Hb_001292_030 |
0.0623115026 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002743_010 |
0.0636244911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002272_210 |
0.0659314952 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000230_420 |
0.0662506128 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 15 |
Hb_005280_020 |
0.0662889241 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 16 |
Hb_001717_060 |
0.0664607093 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 17 |
Hb_002529_120 |
0.0678782036 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 18 |
Hb_005345_020 |
0.0688097473 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 19 |
Hb_001975_120 |
0.0695309004 |
- |
- |
chlorophyll a/b binding protein type II [Glycine max] |
| 20 |
Hb_000512_120 |
0.070002001 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |