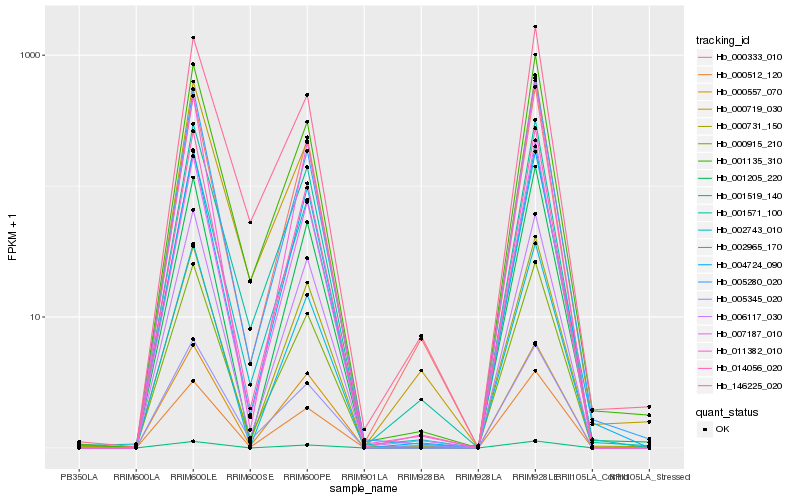
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002743_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_014056_020 |
0.0307285778 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000915_210 |
0.0315518961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001571_100 |
0.0392095147 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 5 |
Hb_001135_310 |
0.0431238527 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 6 |
Hb_000512_120 |
0.0449730014 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 7 |
Hb_000731_150 |
0.0450609635 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 8 |
Hb_006117_030 |
0.0465931699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000557_070 |
0.0467941347 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 10 |
Hb_000333_010 |
0.0490649145 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 11 |
Hb_001205_220 |
0.0494210155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007187_010 |
0.0501810479 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 13 |
Hb_002965_170 |
0.0504738685 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 14 |
Hb_001519_140 |
0.0531962912 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 15 |
Hb_005280_020 |
0.053269455 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 16 |
Hb_000719_030 |
0.0543606764 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_004724_090 |
0.055258054 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 18 |
Hb_011382_010 |
0.0570018314 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 19 |
Hb_146225_020 |
0.0636244911 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 20 |
Hb_005345_020 |
0.0636776402 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |