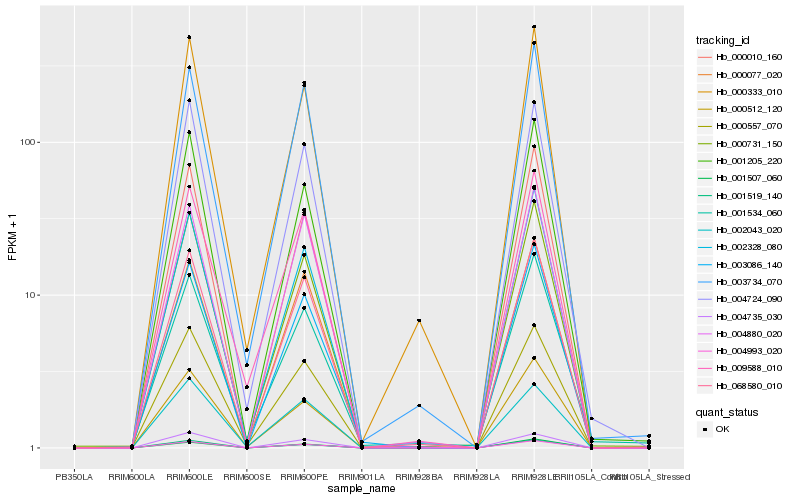
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068580_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 2 |
Hb_004993_020 |
0.0423401593 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_003734_070 |
0.0431927138 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 4 |
Hb_000557_070 |
0.045501839 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 5 |
Hb_000731_150 |
0.0477472299 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 6 |
Hb_000077_020 |
0.0508786094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004880_020 |
0.0530930476 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Jatropha curcas] |
| 8 |
Hb_002328_080 |
0.0548452007 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_003086_140 |
0.056378763 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 6-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_004724_090 |
0.0580340342 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 11 |
Hb_009588_010 |
0.0599253823 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 12 |
Hb_000333_010 |
0.0608590453 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 13 |
Hb_001507_060 |
0.0651064284 |
- |
- |
translation initiation factor eif-2b gamma subunit, putative [Ricinus communis] |
| 14 |
Hb_004735_030 |
0.0670580361 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 15 |
Hb_001519_140 |
0.0683528366 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 16 |
Hb_001205_220 |
0.0685860879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000512_120 |
0.0692962359 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 18 |
Hb_000010_160 |
0.0702874938 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 19 |
Hb_001534_060 |
0.0704730901 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 20 |
Hb_002043_020 |
0.0704779395 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |