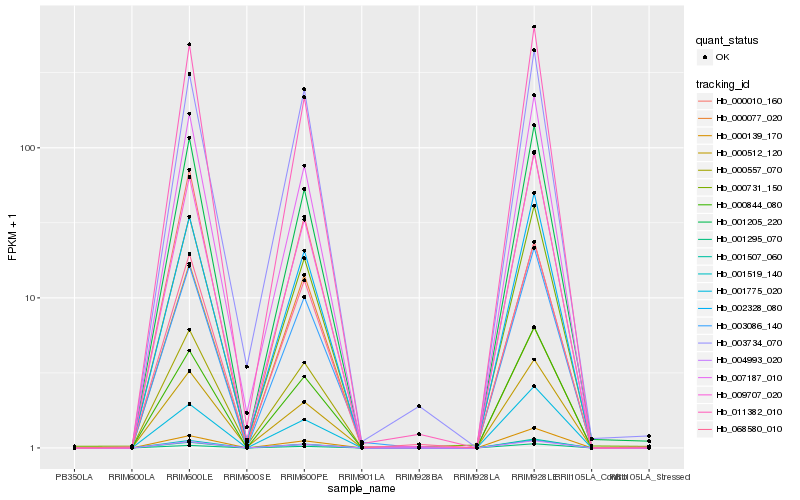
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003086_140 |
0.0 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 6-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_004993_020 |
0.0418950427 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_002328_080 |
0.0440903562 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_001507_060 |
0.0450339421 |
- |
- |
translation initiation factor eif-2b gamma subunit, putative [Ricinus communis] |
| 5 |
Hb_000010_160 |
0.0466559365 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 6 |
Hb_000844_080 |
0.047031645 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 7 |
Hb_009707_020 |
0.0505570472 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 8 |
Hb_000731_150 |
0.0536394787 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 9 |
Hb_001295_070 |
0.0541193076 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 10 |
Hb_001775_020 |
0.0561424886 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 11 |
Hb_068580_010 |
0.056378763 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 12 |
Hb_011382_010 |
0.0569680802 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 13 |
Hb_001205_220 |
0.0570307438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001519_140 |
0.0604064875 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 15 |
Hb_000077_020 |
0.0619040554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007187_010 |
0.0622155165 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 17 |
Hb_000139_170 |
0.0670660326 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 18 |
Hb_003734_070 |
0.0681730377 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 19 |
Hb_000512_120 |
0.0693241406 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 20 |
Hb_000557_070 |
0.0698649962 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |