| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
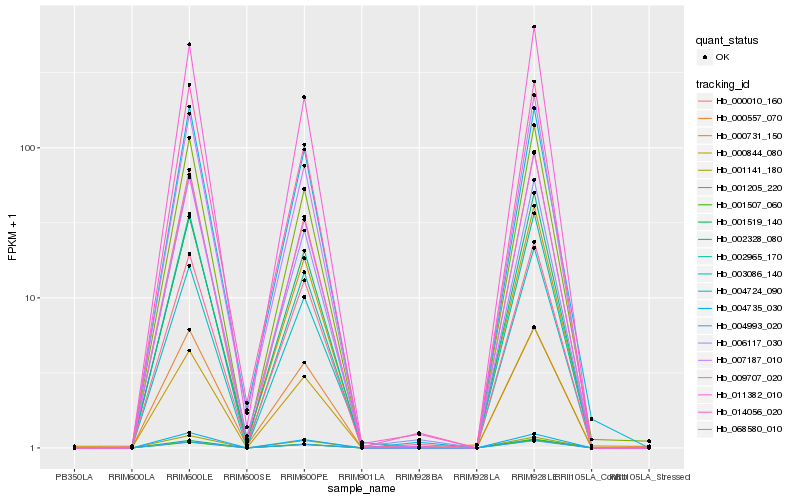
Hb_004993_020 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_001519_140 |
0.0355155352 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Vitis vinifera] |
| 3 |
Hb_003086_140 |
0.0418950427 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 6-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_068580_010 |
0.0423401593 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 5 |
Hb_000731_150 |
0.0437066067 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 6 |
Hb_000557_070 |
0.0449734505 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 7 |
Hb_000010_160 |
0.0453466721 |
- |
- |
Uncharacterized protein TCM_014508 [Theobroma cacao] |
| 8 |
Hb_004735_030 |
0.0461801632 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 9 |
Hb_001205_220 |
0.0473328823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002328_080 |
0.0511313471 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_004724_090 |
0.051338111 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 12 |
Hb_002965_170 |
0.0548514325 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 13 |
Hb_011382_010 |
0.0575027294 |
- |
- |
hypothetical protein CICLE_v10028106mg [Citrus clementina] |
| 14 |
Hb_001507_060 |
0.0591762367 |
- |
- |
translation initiation factor eif-2b gamma subunit, putative [Ricinus communis] |
| 15 |
Hb_014056_020 |
0.0602440576 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000844_080 |
0.060448467 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 17 |
Hb_009707_020 |
0.0606371635 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 18 |
Hb_001141_180 |
0.0617401291 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 19 |
Hb_007187_010 |
0.0621738219 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 20 |
Hb_006117_030 |
0.0624414552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |