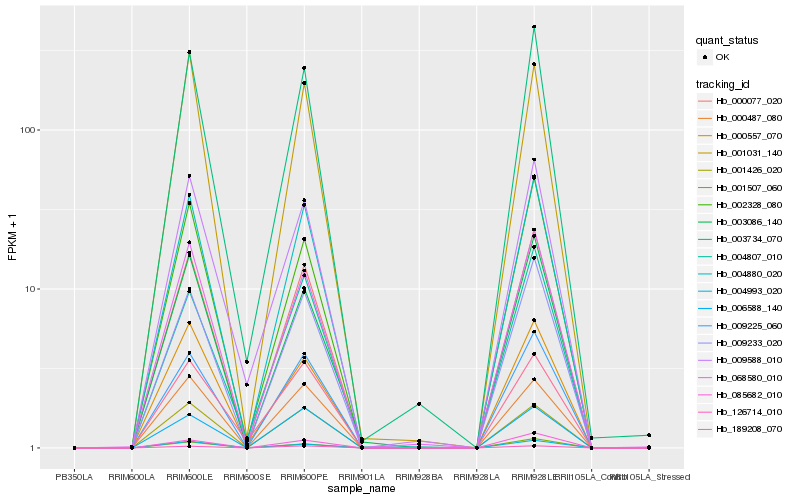
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004880_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Jatropha curcas] |
| 2 |
Hb_000077_020 |
0.0223024135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009225_060 |
0.0313080246 |
- |
- |
PREDICTED: myosin heavy chain kinase B-like [Populus euphratica] |
| 4 |
Hb_003734_070 |
0.0434375526 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 5 |
Hb_009233_020 |
0.0436748941 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 6 |
Hb_068580_010 |
0.0530930476 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 7 |
Hb_001426_020 |
0.0699850331 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 8 |
Hb_004993_020 |
0.0714249711 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_009588_010 |
0.0729458062 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 10 |
Hb_000487_080 |
0.0736501512 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 11 |
Hb_003086_140 |
0.0756862319 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 6-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001507_060 |
0.077564645 |
- |
- |
translation initiation factor eif-2b gamma subunit, putative [Ricinus communis] |
| 13 |
Hb_126714_010 |
0.0776770382 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 14 |
Hb_006588_140 |
0.0780921392 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_085682_010 |
0.0798812775 |
- |
- |
- |
| 16 |
Hb_001031_140 |
0.082038216 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 17 |
Hb_002328_080 |
0.0828184302 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000557_070 |
0.0832707988 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 19 |
Hb_004807_010 |
0.0844289774 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 20 |
Hb_189208_070 |
0.0845957878 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |